Team:Czech Republic/Project/Location tags
Location Tags
Contents
Abstract
To guarantee the ability of our system to recognize, bind, and respond to the presence of a tumour cell, we selected five antibody fragments to display on our yeast cells using the yeast display technology and monitor the cells' binding. Their expression was detected and monitored on a microfluidic chip and was also visualized with the use of antibody staining and fluorescence microscopy.
Key Achievements
- Expressed streptavidin, EpCAM, Anti-EpCAM scFv, c-Myc scFv and anti-HuA scFv on the surface of our yeast cells
- Monitored the dynamic binding of the displayed fragments of antibodies and corresponding markers
- Demonstrated compatibility of our yeast system with blood and functionality of the location tags in blood plasma
Introduction
To enable our modified Saccharomyces cerevisiae cells to bind extracellular markers, we expressed single-chain variable fragments of antibodies (scFvs) on their surface. These proteins are fused with the adhesion subunit of a-agglutinin, which is a protein naturally used by S. cerevisiae for aggregation during mating. Binding of the antibody to its specific marker enables full development of a signal transduction dependent on the pheromone signaling pathway. We proposed a modified yeast display technique to enable our cells to present the antibody fragments on their surface at all times. We were able to detect and measure the binding of several oncogenic, epithelial, and blood markers.
Agglutinins
Alpha-agglutinin and a-agglutinin are cell wall glycoproteins of S. cerevisiae that possess complementary surfaces and mediate cell-to-cell adhesion during mating [Lipke and Kurjan, 1992]. A-agglutinin consists of two subunits, of which Aga1p is anchored to the membrane by a glycosylphosphatidylinositol anchor (GPI) and to which the second subunit, Aga2p, is bound by two disulphide bonds. Alpha-agglutinin is composed of only one subunit and is connected to a modified GPI anchor. They are both highly glycosylated and constitutively expressed in low levels [Zhao et al, 2001].
Yeast Display
The Yeast Surface Display was first described in 1997 by E. Border and K. Wittrup as a method for engineering cell surface and secretion proteins. It provides a great advantage of the possibility of post-translational modifications in a eukaryotic organism - yeast. One of the most widespread uses of Yeast Display is screening polypeptide libraries for high-affinity antibodies[Boder and Wittrup, 1997].
Design
Traditional Yeast Display
The gene for the protein of interest to be displayed on a cell's surface is fused in frame to the AGA2 gene on a yeast display plasmid. The second part of the display, AGA1 gene is integrated into a chromosome in a special S. cerevisiae strain EBY100. Both of the constructs are originally under a GAL1 promoter, thus requiring galactose to be present in the medium for the system to function. [Baird et al, 2012].
We obtained the original yeast display system from professor Sheldon Park, which we are thankful for. This particular yeast display plasmid used the BamHI and NheI restriction sites for insertion of scFv sequences and possessed a hemagglutinin tag upstream of the scFv sequence insertion site and a FLAG epitope at the downstream end.
Choice of Markers
To develop and test our system we chose to express on the surface of our S. cerevisiae cells the following markers.
Biotin
Biotin was our first choice of a marker because of its extremely strong bond with streptavidin. It could give us not only great results but also a benchmark affinity value to compare the other antibodies’ affinities to.
EpCAM
EpCAM, the transmembrane epithelial cell adhesion molecule, functions in cell signaling, proliferation, differentiation, and is present in many carcinomas. We selected it because of its significance in tumor growth promotion and its ability to bind Anti-EpCAM, which we also expressed on S. cerevisiae using yeast display.
c-Myc
C-Myc is a transcription factor that regulates cell growth, apoptosis, and cellular differentiation. Its levels are often upregulated in different cancer types, where it plays a role in gene duplication and Ras interaction.
HuA
HuA stands for human antigen A of red blood cells. The decision to enable the expression of an antibody against antigen A on yeast surface allows us to test our system directly in blood and prove that the positive agglutination reaction is specific and clearly visible.
Modified Yeast Display
Furthermore, we made several modifications to the yeast display method in order for it to better suit our needs. The goal was to simplify it and reduce the amount of new DNA that needs to be introduced into a yeast cell. The first of two alternatives was based on creating a general cassette containing a promoter, a terminator, and necessary cloning sites. The second alternative meant attaching the specific antibody with a terminal epitope tag directly to Agα1 protein.
Expression Cassette
We wanted to build five variations of the pRSII 413 plasmid based on which final cassette will act as the insert. The design of our cassette included using the constitutive pTv3 promoter, and a terminator sequence. One of the five final cassettes would contain the sequence coding for the Aga1 protein, and four of the cassette inserts would contain sequences coding for Aga2 protein fused with a single chain antibody of choice. By carefully designed consequent PCR reactions and restriction digests we aimed to obtain in vitro the 1) promoter, 2) terminator, 3) promoter and terminator, 4) promoter, ORF and terminator. Subsequent restriction digests and ligations would lead to a complete expression plasmid containing our desired inserts.
Agα1p version
By fusing the antibody fragment directly to the Agα1 protein, we are able to eliminate the complications that arise because of the translation of several proteins in a row. The courage to pursue this alternative stemmed from the positive results obtained by J. Wasilenko and colleagues in their work of displaying the hemagglutinin of avian influenza virus on yeast cells using alpha-agglutinin. We aimed to incorporate into only one plasmid the sequence for an antibody fused with the C-terminal end of Agalpha1 (alpha-agglutinin) and a secretion signal. All of it to be controlled by one promoter [Wasilenko et al, 2010]. This alternative greatly appealed to us thanks to its elegance and simplicity but proved to be quite time-consuming to prepare. Therefore, we only finished the design of the system and assigned the idea for future experiments.
Construction
Construction of the Traditional Yeast Display Plasmids
We used modified pCT302 plasmid (https://www.addgene.org/41845/) and inserts described below for the construction of our final yeast display plasmids. Strain EBY100 was used for yeast display experiments. Expression of the protein display on the yeast surface can be readily induced by addition of galactose to the growing medium.
More information about sequences used in yeast display experiments:
EpCAM
EpCAM protein consists of 314 amino acids. We used the sequence encoding only the extracellular part of the EpCAM protein (729 bps long). We also added restriction sites for BamHI, NheI (yeast display) and SpeI, EcoRI (EpCAM production) endonucleases with PCRs.
ATGCAAGAAGAATGTGTATGTGAAAATTATAAACTAGCTGTCAACTGTTTTGTAAATAACAATAGACAGTGTCAATGTACTTCCGTTGGCGCTCAAAACACAGTTATCTGTTCTAAACTAGCCGCGAAGTGTCTTGTTATGAAGGCAGAGATGAATGGCTCCAAACTGGGTAGGCGTGCCAAGCCTGAGGGTGCCCTACAGAACAATGATGGGCTTTACGATCCAGATTGTGATGAATCCGGTTTATTCAAAGCCAAGCAGTGTAATGGTACTTCCATGTGCTGGTGTGTAAACACCGCTGGAGTGAGGAGAACCGATAAAGACACGGAAATCACCTGTAGCGAAAGAGTCAGGACTTACTGGATCATTATCGAACTGAAGCACAAAGCTAGGGAGAAGCCTTATGACAGCAAAAGTTTGAGGACTGCCTTACAGAAAGAAATCACAACTAGGTATCAATTAGACCCGAAATTTATAACGAGTATCCTTTACGAGAATAACGTCATTACCATTGATCTAGTGCAAAATTCTTCCCAGAAGACTCAAAATGACGTTGATATTGCAGATGTTGCCTATTACTTTGAAAAAGATGTGAAAGGAGAGAGTTTGTTCCACTCAAAAAAGATGGATCTGACAGTTAACGGGGAACAGTTAGATCTTGATCCGGGTCAAACGTTAATATATTATGTTGACGAAAAAGCACCAGAATTTAGTATGCAGGGATTGAAATAA
The sequence was extracted from the Uniprot database and further optimized for expression in yeast (http://www.uniprot.org/uniprot/P16422).
Anti-c-Myc (9E10)
This sequence encodes part of the 9E10 Anti-cMyc antibody (scFv) and is 780 bps long. It consists of sequences encoding light and heavy chains of the antibody. These sequences are connected by a Yol linker containing a Yol epitope. Restriction sites recognized by BamHI and NheI were added for easy cloning into the yeast display plasmid.
ATGCAGGTGCAGCTaCAGGAGTCTGGGGGAGACTTAGTGAAGCCTGGAGGGTCCCTGAAACTCTCCTGTGCAGCCTCTGGATTCACTTTCAGTCACTATGGCATGTCTTGGGTTCGCCAGACTCCAGACAAGAGGCTGGAGTGGGTCGCAACCATTGGTAGTCGTGGTACTTACACCCACTATCCAGACAGTGTGAAGGGACGATTCACCATCTCCAGAGACAATGACAAGAACGCCCTGTACCTGCAAATGAACAGTCTGAAGTCTGAAGACACAGCCATGTATTACTGTGCAAGAAGAAGTGAATTTTATTACTACGGTAATACCTACTATTACTCTGCTATGGACTACTGGGGTCAAGGAGCCTCAGTCACCGTCTCCTCAGCCAAAACAACACCCAAGCTTGAAGAAGGTGAATTTTCAGAAGCACGCGTAGATATCGTTCTCACTCAATCTCCAGCTTTCTTGGCTGTATCTCTAGGACAGAGGGCCACCATCTCCTGtAGAGCCAGCGAAAGTGTTGATAATTATGGCTTTAGTTTTATGAACTGGTTCCAACAGAAACCAGGACAGCCACCCAAACTCCTCATCTATGCTATATCCAACCGAggttctGGGGTCCCTGCCAGGTTTAGTGGCAGTGGGTCTGGGACAGACTTCAGCCTCAACATCCATCCTGTAGAGGAGGATGATCCTGCAATGTATTTCTGTCAGCAAACTAAGGAGGTTCCGTGGACGTTCGGTGGAGGCACCAAGCTGGAAATCAAACGGGCTGTTGCTTAA
The sequence was extracted from the article "Primary structure and functional scFv antibody expression of an antibody against the human protooncogene c-myc." and further optimized for expression in yeast (http://www.ncbi.nlm.nih.gov/pubmed/9219032).
Anti-EpCAM
This sequence encodes part of the C215 Anti-EpCAM antibody (scFv) and is 756 bps long. It consists of sequences encoding light and heavy chains of the antibody. These sequences are connected through a G4S linker. Restriction sites recognized by BamHI and NheI were added for easy cloning into the yeast display plasmid.
ATGCAGGTCAAGCTtCAGCAGTCAGGGGCTGAACTGGTGAGGCCTGGGGCTTCAGTGAAGCTGTCCTGCAAGGCTTCTGGCTACACCTTCACCAACTACTGGATAAACTGGGTGAAGCAGAGGCCTGGACAAGGCCTTGAGTGGATCGGAAATATTTATCCTTCTTATATTTATACTAACTACAATCAAGAGTTCAAGGACAAGGTCACATTGACTGTAGACGAATCCTCCAGCACAGCCTACATGCAGCTCAGCAGCCCGACATCTGAGGACTCTGCGGTCTATTACTGTACAAGATCCCCTTATGGTTACGACGAGTATGGTCTGGACTACTGGGGCCAAGGCACCACGGTCACCGTCTCCTCAGGTGGAGGCGGTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGGACATCGAGCTCACTCAGTCTCCATCCTCCCTGACTGTGACAGCAGGAGAGAAGGTCACTATGAACTGCAAGTCCAGTCAGAGTCTGTTAAACAGTAGAAATCAAAAGAACTACTTGACCTGGTACCAGCAGAAACCAGGGCAGCCTCCTAAACTGTTGATATACTGGGCATCCACTAGGGAATCTGGGGTCCCTGATCGCTTCACAGGCAGTGGATCTGGAACAGATTTCACTCTCACCATCAGCAGTGTGCAGGCTGAAGACCTGGCAGTTTATTACTGTCAGAATGATTATGTTTATCCGCTCACGTTCGGTGCTGGGACCAAGCTGGAAATAAAACGGTAA
The sequence was extracted from the European Nucleotide Archive database and further optimized for expression in yeast (http://www.ebi.ac.uk/ena/data/view/AJ564232).
Anti-HuA
This sequence encodes part of the AC-1001 Anti-HuA (Human antigen A) antibody (scFv) and is 735 bps long. It consists of sequences encoding light and heavy chains of the antibody. These sequences are connected by a G4S linker. Restriction sites recognized by BamHI and NheI were added for easy cloning into the yeast display plasmid.
ATGCAAGTCCATTTAGTGGAGTCAGGTGGTGGCGTAGTTCAACCTGGAAGATCATTGACACTTTCCTGTGCTGCGTCCGGTTTTACCTTCAATTCTTATACGTTCCACTGGGTAAGACAAACCCCAGGTAAAGGTCTTGAATGGGTTGCAGTTTTAGCATACGACGGGAGTTACCAGCATTATGCAGATTCCGTGAAAGGAAGATTTACAATTTCCAGAGATAACAGCAAAAATACTTTATACTTGCAAATGAATAGTCTGAGACTTGAAGACACAGCGGTATACTTCTGTGCAAGGGGACAGACTACAGTTACTAAAATTGATGAGGACTATTGGGGGCAGGGTACATTGGTAATTGTCTCCTCCGGTGGTGGCGGTAGTGGAGGCGGTGGCTCCGGGGGAGGGGGCTCTGAGATCGTCCTTACCCAGTCTCCAGCAACGCTTAGTTTGTCTCCTGGAGAAAGAGCTACACTTTCTTGCAGAGCAAGCCAGTCTGTCTCTTCTTACTTAGCTTGGTATCAGCAAAAACCAGGTCAGGCTCCAAGGTTGCTGATATATGATGCATCTAACAGGGCGACTGGTATTCCTGCCAGGTTTTCCGGCTCCGGTTCTGGCACTGACTTCACACTTACTATTTCATCTCTAGAACCAGAAGATTTTGCCGTTTATTACTGCCAGCAGAGATCTAATTGGCCTAGGTCTTTTGGTCAGGGTACGAAGGTCGAGATCAAAAGGTAA
The sequence was extracted from the article "Human and mouse monoclonal antibodies to blood group A substance, which are nearly identical immunochemically, use radically different primary sequences." and further optimized for expression in yeast (http://www.ncbi.nlm.nih.gov/pubmed/7759488).
Streptavidin
The gene encoding streptavidin protein is 381 bps long. Sequences containing priming sites for M13 forward and reverse primers were added at the ends of the synthesized streptavidin so that streptavidin gene could be amplified with these commonly used primers. Cutting sites for BamHI and NheI restriction endonucleases were also added for easy cloning into the yeast display plasmid.
ATGGCTGAAGCTGGTATCACCGGCACCTGGTACAACCAGCTGGGTTCCACCTTCATCGTTACCGCTGGTGCTGACGGTGCTCTGACCGGTACCTACGAATCCGCTGTTGGTAACGCTGAAAGCCGCTACGTTCTGACCGGTCGTTACGACTCCGCTCCGGCTACCGACGGTTCCGGAACCGCTCTGGGTTGGACCGTTGCTTGGAAAAACAACTACCGTAACGCTCACTCCGCTACCACCTGGTCTGGCCAGTACGTTGGTGGTGCTGAAGCTCGTATCAACACCCAGTGGTTGTTGACCTCCGGCACCACCGAAGCCAACGCGTGGAAATCCACCCTGGTTGGTCACGACACCTTCACCAAAGTTAAACCGTCCGCTGCTTCTTAA
Sequence was extracted from the iGEM parts database and further optimized for expression in yeast (http://parts.igem.org/Part:BBa_K283010).
Construction of the Expression Cassette
Sequence of the Aga1 protein was amplified from yeast genome using Q5 polymerase and sequences of the pTv3 promoter and CYC terminator was amplified from plasmids provided by Module 1 (working on synthetic haploids). The primers we used are disclosed in the table below.
| Name | Sequence |
|---|---|
| pTv3 down | TTAACAGAATTCGGATTCTCACAATCCTGTCGGT |
| pTv3 up | ATAATTCTCGAGAAGCTTGGACTTCCCACCGCCTTC |
| CYC TT down | ACAAAAGTCGACTCTAGACGAGCGTCCCAAAACCT |
| CYC TT up | ATATTAGAATTCATTATTAAATGGATCCTCATGTAATTAGTTATGTCACGCT |
| Aga1 down | ATATATGGATCCTTAACTGAAAATTACATTGCAAGCA |
| Aga1 up | ATATTAGAATTCACCACCATGACATTATCTTTCGCTCATTTTAC |
The amplification of Aga1 and assembly of the promoter and the terminator in one sequence was successful as proved by gel electrophoresis and subsequent gel purification. However, even after many repeated tries to clone the premature cassette into the pRS413 and pRS415 plasmids, our efforts remained futile. We tried using different ligases, amount of electroporated cells, and even a different cloning vector. As a result, we turned our focus solely on testing and improving the first of the three approaches - the original yeast display system.
Materials and Methods
Strains
E. coli
- DH5α
S. cerevisiae
- EBY100
- BY4741
- 7284 MATa
Materials
- LB-M agar plates with ampicillin
- 1.5 ml eppendorf tubes
- 0.5 ml PCR tubes
- 50 ml centrifuge tubes
- NucleoSpin Plasmid DNA, RNA, and protein purification Kit
- NucleoSpin Gel and PCR Clean-up Kit
- LB-M medium with chloramphenicol
- YPD medium
- SD-min medium + CAA
- NaOH agarose gel and buffer
- TAE agarose gel and buffer
Methods
- Transformation
- Minipreparation of plasmid DNA
- Restriction digest
- Ligation
- NucleoSpin gel clean-up
- NucleoSpin plasmid DNA purification
- Induction of protein display
- Fluorescent immunostaining
Detailed description of all methods can be found on our Protocols page.
Results
Validation of displayed proteins by immunofluorescent labeling
Successful surface display of our proteins was confirmed by immunofluorescent staining. Yeast display plasmid enables labeling of the fused protein by anti-HA and anti-FLAG antibodies either fused directly to a fluorophore or subsequently tagged by a corresponding fluorophore-marked secondary antibody. We have stained our modified yeast strains with both anti-HA and anti-FLAG antibodies. Presence of the fused proteins on the surface can be distinguished by a visible "glowing corona". Pictures were made by merging phase field images and images showing the fluorescent signal. Pictures showed that the majority of the yeast cells successfully displayed the proteins, there were however some cells which displayed the protein in lower amounts or did not display any Aga2-fused proteins at all.
Verification of interactions between EpCAM and Anti-EpCAM scFv
Interaction of the cancer marker EpCAM and Anti-EpCAM scFv was harder to prove. Yeast display system should be induced in 20°C, as yeast cells might have difficulties with assembly of some foreign proteins in higher temperatures. Lower temperatures should slow down the rate of protein assembly and secure correct folding of the expressed proteins. However, induction in these conditions led to clumping of the yeast cells expressing both the Anti-EpCAM scFv and the EpCAM protein. We therefore decided to examine the binding strength under various conditions.
Samples extracted from both cultures were vortexed and incubated for a brief period of time. The difference between sizes and frequency of cell aggregates in the negative control (only EpCAM or Anti-EpCAM cells) and positive controls (mixed EpCAM and Anti-EpCAM displaying cells) was visible at first glance. Inspection of the cell aggregates under the microscope revealed enormous differences between these two samples. Occurrence of this phenomenon could be explained by the affinity of anti-EpCAM antibody on the surface of the first type of cells for displayed EpCAM proteins on the surface of the second type of cells. As conditions which accompanied this experiment were not optimal, these results cannot be upscaled to full-scale conclusions. They nevertheless indicate some difference in the ability of marker and its receptor to bind each other.
This experiment should simulate the real situation in IOD-mediated CTC detection. Individual yeast cells displaying EpCAM protein represented circulating tumor cells, whereas Anti-EpCAM displaying cells represented the first degree of detection.
Yeast-induced blood agglutination
One of the biggest achievements of this module was successful yeast-mediated induction of blood agglutination. Yeast cells (transformed EBY100 cells) used in this experiment were displaying scFv against the Human Antigen A (HuA). Yeast cells that served as a negative control were the exactly same cells (EBY100), but without anti-HuA display plasmid. Donor of the blood had A+ blood group. Our modified yeast cells could therefore interact with the antigen A on the surface of the red blood cells. The experiment proved capability of the modified yeast cells to agglutinate blood. This reaction should be at the very end of the signal transduction pathway and formation of blood clumps in a blood sample should indicate the presence of the particular cancer cells.
The master experiment
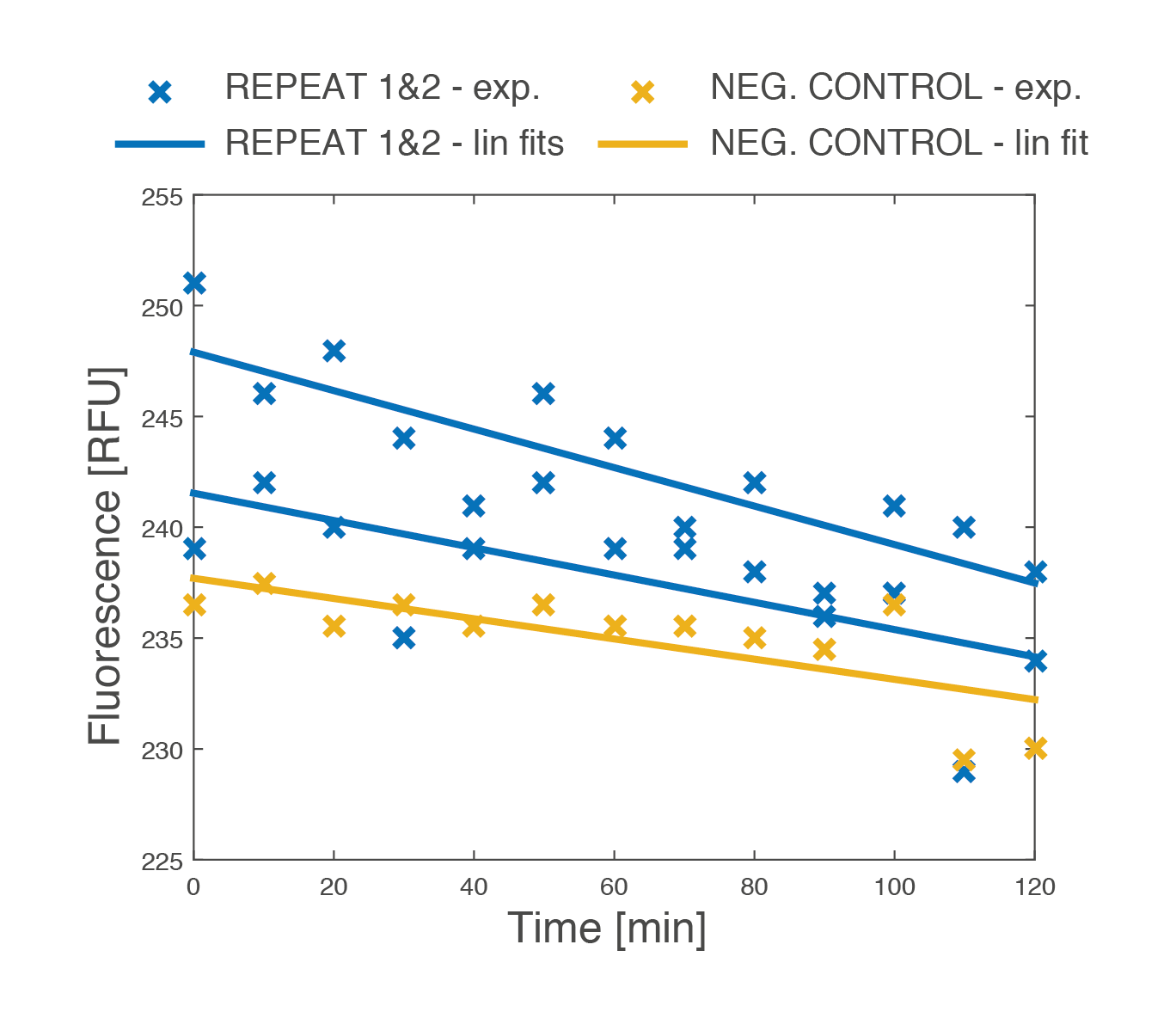
Influence of the relative position of the yeast cells to each other on the pheromone pathway signal transduction was examined in our last experiment. Two different yeast strains were used. The first type displayed Anti-EpCAM scFvs on its surface and also produced wild-type mating pheromones after the induction with copper sulfate solution. The second type displayed EpCAM proteins on its surface and also had a reporter plasmid that could react to concentrations of wild-type mating pheromone in the environment by GFP production. In other words, fluorescence was dependent on the concentration of wild-type mating pheromone in the proximity of the reporter yeast cell.
This experiment confirmed that the fluorescence of the reporter type yeast cells is indeed dependent on the activation with a mating pheromone. It also suggested that clumping dependent on the expression of complementary proteins helps this activation. Yeast cells with a reciprocal affinity for each other should stay close, and thus the frequency and intensity of the signal should increase. We observed this exact mechanism that was dependent on the display of complementary proteins on the yeast cell surface. Yeast cells that were unable to bind each other showed a decreased intensity of fluorescence.
References
- ↑ Zhao, H.,Shen, Z.-M., Kahn, P. C. and Lipke, P. N. (2001). Interaction of α-Agglutinin and a-Agglutinin, Saccharomyces cerevisiae Sexual Cell Adhesion Molecules. Journal of Bacteriology, 183(9), 2874–2880. doi:10.1128/JB.183.9.2874-2880.2001
- ↑ Lipke, P. N. and Kurjan, J. (1992). Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiology and Molecular Biology Reviews, 56(1), 180-194.
- ↑ Boder, E. T. and Wittrup, K. D. (1997). Yeast surface display for screening combinatorial polypeptide libraries. Nature Biotechnology, 15, 553-557. doi:10.1038/nbt0697-553
- ↑ Baird, C. et al. (2012). Yeast Display scFv Antibody Library User’s Manual. Richland, WA: Pacific Northwest National Laboratory.
- ↑ Wasilenko, J. L. et al. (2010). Cell Surface Display of Highly Pathogenic Avian Influenza Virus Hemagglutinin on the Surface of Pichia Pastoris Cells Using alpha-Agglutinin for Production of Oral Vaccines. Biotechnology Progress, 26(2), 542-547. doi:10.1002/btpr.343.