Team:Paris Saclay/Experiments
Labwork
Protocols
- Transformation of super competent E. coli cells
- Mac Conkey Medium Preparation
- Extraction of the Plasmid DNA from Bacteria by using NucleoSpin Plasmid®
- Gel Electrophoresis
- Purification on agarose gel
- Gibson assembly
- Isothermal assembly
Transformation of super competent E. coli cells
Thaw supercompetent cells at room temperature. (almost 100µL for control and 100 µL for one DNA transformation).
- Control
In a 1.5 ml microcentrifuge tube (eppendorf), add 100 μl solution of supercompetent cells (0).
- Plasmid DNA
- In a 1.5 ml eppendorf, add 100 μl solution of supercompetent cells, 1 μl (concentration 0.1 ng/μl) of DNA plasmid (With resistance to an antibiotic)(1).
- Incubate solutions on ice for 30 min.
- Make a heat shock: incubate bacteria at 42 °C for 2min30, then on ice for 2 min.
- Add 900 μl LB in your eppendorfs, incubate solutions with agitation at 37 °C for 1 h.
- Take 100 μl of each solution : the control(0)and the transformed bacteria (1),(2),(3) spread them out on LB plate with the appropriated antibiotic.
- Centrifuge 30s at 11,000*g. Discard 750μl and mix the other 150μl always present in the eppendorf. spread them out on an other plate (always with the appropriated antibiotic).
Mac Conkey Medium Preparation
- In a 1L glass bottle add 500mL of sterile water.
- Add 20g of Mac Conkey (without carbone source)
- Add 5g of carbone source (in our case Sorbitol or Lactose)
- Microwave for 10 to 15min (shake gently each minute)
- Let it cool before adding antibiotic.
- Make your plates under sterile environment.
Extraction of the Plasmid DNA from Bacteria by using NucleoSpin Plasmid®
The experiment is performed with buffer solutions of NucleoSpin® Tissue, protocol 5.1 for bacteria.
- Cultivate and harvest bacterial cells:
Take 2 ml of bacteria culture in a 2 ml microcentrifuge tube (eppendorf), prepare one sample. Centrifuge the culture for 30s at 11,000*g. Discard supernatant and remove as much of the liquid as possible.
- Cell lysis:
Resuspend and dissolve the pellet in 250 µl Buffer A1 by pipetting up and down. Make sure no cell clumps remain before addition of Buffer A2. Add 250 µl Buffer A2. Mix gently by inverting the tube 6-8 times. Do not vortex to avoid shearing of genomic DNA. Incubate at room temperature for up to 5 min or until lysate appears clear. Add 300µl Buffer A3. Mix thoroughly by inverting the tube 6-8 times. Do not vortex to avoid shearing of genomic DNA!
- Clarification of lysate:
Centrifuge for 5 min at 11.000*g at room temperature. Repeat this step in case the supernatant is not clear!
- Bind DNA:
Place a NucleoSpin® Plasmid Column into a collection tube (2ml) and decant the supernatant from step 3 or pipette a maximum of 750µl of the supernatant onto the column. Centrifuge for 1 min at 11.000*g. Discard the flow-through and place the column back into the collection tube. Repeat this step to load the remaining lysate.
- Wash silica membrane:
Add 600 µl Buffer A4 (supplemented with ethanol) to the column and centrifuge for 1 min at 11.000*g. Discard flow-through and place the NucleoSpin Column back into the empty collection tube.
- Dry silica membrane:
Centrifuge the column for 2 min at 11.000*g and discard the collection tube.
- Elute DNA:
Place the NucleoSpin Plasmid Column in a 1.5 ml microcentrifuge tube (not provided), and add 50 µl Buffer AE. Incubate at room temperature for 1 min. Centrifuge for 1 min at 11.000*g.
- Discard the column.
- Analysis:
Check the presence of DNA and determine its concentration by using quantification on electrophoresis gel.
Gel Electrophoresis
Preparing 1% agarose gel:
- Weight out 1g of Agarose powder and add it to a 500 ml flask
- Add 100 ml of TAE Buffer to the flask.
- Melt the agarose in a microwave until the solution become clear (heat the solution for several short intervals - do not let the solution boil for long periods as it may boil out of the flask).
- Let the solution cool to about 50-55°C, swirling the flask occasionally to cool it evenly.
- Add BET with a final concentration of 200ng/µL.
- Place the combs in the gel casting tray.
- Pour the melted agarose solution into the casting tray and let cool until it is solid (it should appear milky white).
- Carefully pull out the combs and place the gel in the electrophoresis chamber.
Loading Samples and Running an Agarose Gel:
- Place the gel in the electrophoresis chamber.
- Add enough TAE Buffer 1X so that there is about 2-3 mm of buffer over the gel.
- Add loading buffer 6X to each of your samples.
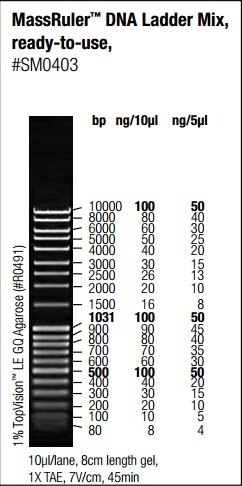
- Carefully load a molecular weight ladder into the first lane of the gel.
- Carefully load your samples into the additional wells of the gel.
- Record the order each sample will be loaded on the gel, including who prepared the sample, the DNA template - what organism the DNA came from, controls and ladder.
- Run the gel usually at 80V during 30 mins or until the dye line is approximately 75-80% of the way down the gel.
- Turn OFF power, disconnect the electrodes from the power source, and then carefully remove the gel from the gel box.
- Using any device that has UV light, visualize your DNA fragments.
Purification on agarose gel
Collect of DNA Samples:
- Weigh empty eppendorfs (they will be used to collect gel extracts).
- Using any device that has UV light, visualize your DNA fragments.
- Cut with a scalpel each strip visualized on UV light and collect them in one of empty eppendorf.
Melting Agarose Gel:
- Put eppendorfs containing gel in the heating bloc adjusted at 56°C for 20min (or when all the agarose gel is melted)
- Pursue the protocol by the NucleoSpin® Gel and PCR Clean-up protocol
Gibson assembly
1. Principle: One-step isothermal in vitro recombination
Two adjacent DNA fragments sharing terminal sequence 20 bp overlaps were joined into a covalently sealed molecule in a one-step isothermal reaction.
T5 exonuclease removed nucleotides from the 5' ends of doublestranded DNA molecules, complementary single-stranded DNA overhangs annealed.
Phusion DNA polymerase filled the gaps and Taq DNA ligase sealed the nicks.T5 exonuclease is heat-labile and is inactivated during the 50°C incubation.
Incubation at 50°C during 1 hour --> the product can be used to transform into E. coli directly.
2. Design primer
Primer overlap at least 20 bp with the adjacent fragments (plasmid, fragments)
Order primer: IDT CompanyScale: 25nm DNA oligoNormalization: NonePurification: standard desalting
The Tm is defined as the temperature in degrees Celsius, at which 50% of all molecules of a given DNA sequence are hybridized into a double strand, and 50% are present as single strands (the melting temperature).Tm of overlap part must be higher than 50°C.
3. PCR amplification
Using enzyme Phusion High-Fidelity DNA polymerase (the most accurate thermostable polymerase available).
4. Clean PCR, plasmid
- For DNA fragments: Gel purify if it has an unspecific band or purify PCR product by kit. Estimate the concentration.
- For plasmid: after PCR, digest your PCR product by enzyme DpnI (recognize and cut methylated DNA, allow to remove remaining plasmid template, and does not cleave PCR-amplified DNA). Then, purify your PCR plasmid digestion and estimates the concentration.
5. Gibson amplification
- Add the combined fragments (5 µl) to one Isothermal Assembly reaction aliquot (15 µl) and mix by pipetting (20 µl total).
- Not exceeding a total volume of 20 µl. If required, bring to 5 µl with ddH2O. 100 ng of plasmid is recommended.
- Incubate the mix at 50°C for 60 min.
- Transform E. coli DH5α with 10 µl of assembly mix.
6. Transformation in E. coli
Negative control: transform E. coli with the plasmid PCR product digested with DpnI --> know the background.
Isothermal assembly
Isothermal Assembly works by combining a cocktail of exonuclease, polymerase, and ligase to fuse dsDNA fragments with sufficiently (20-120 bp) homologous ends. It leaves no "scar" behind, i.e. you can expect your product to contain the EXACT overlap sequence. The reaction may work with shorter ends (e.g. 15 bp), so long as the annealing temperature is higher than 50C.
To perform isothermal assembly:
- PCR up your fragments of choice, and purify. Also gel purify the cut vector.
- Not exceeding a total volume of 5 µl, in a PCR tube, combine fragments at equal molecular ratio. If required, bring to 5 µl with ddH2O. Use approx. 100 ng of plasmid backbone.
- Add the combined fragments (5 µl) to 1 Isothermal Assembly reaction aliquot (15 µl) and mix by pipetting (20 µl total).
- Place mix at 50°C for 60 min.
- Transform with 10 µl of assembly mix.
References:
Gibson et al (2009) Nature Methods 6(5):343-345.
Making Isothermal Assembly Aliquots
5x isothermal assembly reaction buffer (assemble on ice):
| From the paper: | Actually added: |
| 3 mL 1M Tris-HCl pH 7.5 | 3 mL 1M Tris-HCl pH 7.5 |
| 150 µL 2M MgCl2 | 300 µL 1M MgCl2 |
| 60 µL 100 mM dGTP | 600 µL 10 mM each dNTP |
| 60 µL 100 mM dCTP | |
| 60 µL 100 mM dTTP | |
| 60 µL 100 mM dATP | |
| 300 µL 1M DTT/td> | 300 µL 1M DTT |
| 1.5 g PEG-8000 | 1.5 g PEG-8000 |
| 300 µL 100 mM NAD | 20 mg NAD |
| ddH2O to 6 ml | ddH2O to 6 ml |
Prepare 320 µL aliquots (18) and freeze all but one. * Label these “5X isotherm buffer”To the one remaining (320 µL), add:
|
1.2 µL |
T5 Exonuclease |
|
20 µL |
Phusion polymerase |
|
160 µL |
Taq ligase |
|
700 µL |
ddH2O |
Prepare 15 µL aliquots (~80) on ice in PCR tubes and store at -20C. These should be good for up to a year.
Buffer
|
Item |
Vendor |
Ref |
|
1M Tris-HCl pH 7.5 |
Sigma |
T2319 |
|
Magnesium Chloride, 1.00 +/- 0.01M Solution |
Affymetrix / USB |
78641 10 x 1 ml |
|
Nicotinamide adenine dinucleotide (NAD) |
VWR |
A1124,0005 |
|
DTT, molecular biology grade |
FERMENTAS |
R0861 |
|
Polyethylene Glycol 8000, Powder |
USB / Affymetrix |
19966 |
|
dNTP Mix, 10mM each |
Fermentas |
R0192 1 ml |
Enzymes:
|
T5 Exonuclease |
NEB |
M0363S |
|
Taq DNA Ligase |
NEB |
M0208L |
|
Phusion™ High-Fidelity DNA Polymerase |
Thermoscientific |
F530-S |