Team:UESTC Software/Collaborations
Collaborations
Photo From © PEXELS
Help SCU_China by developing SD Finder
In our communication activities, SCU_China team introduced to us an interesting idea. They want to find out SD sequence in DNA and analyze the strength of the SD sequence that affects translation issues. Then, they can standardize the strong SD sequence, so that it can be reused in future.
But there are two barriers in their works. One is how to find out all of unknown SD sequence in a DNA sequence. The other one is how to analyze the strength of SD sequence.
To deal with these two problems, we cooperated with SCU_China team(https://2015.igem.org/Team:SCU_China/Collaborations) and CHINA_CD_UESTC(https://2015.igem.org/Team:CHINA_CD_UESTC/Collaborations) team to study how to find out SD sequence and analyze the strength of it.
Then we developed an online software based on browser—SD Finder, which is used in helping users predict the location of ORF and SD sequence in DNA. User only needs to type sequence that needs to be predicted, select the initiation codons and termination codons. Then comes the prediction.
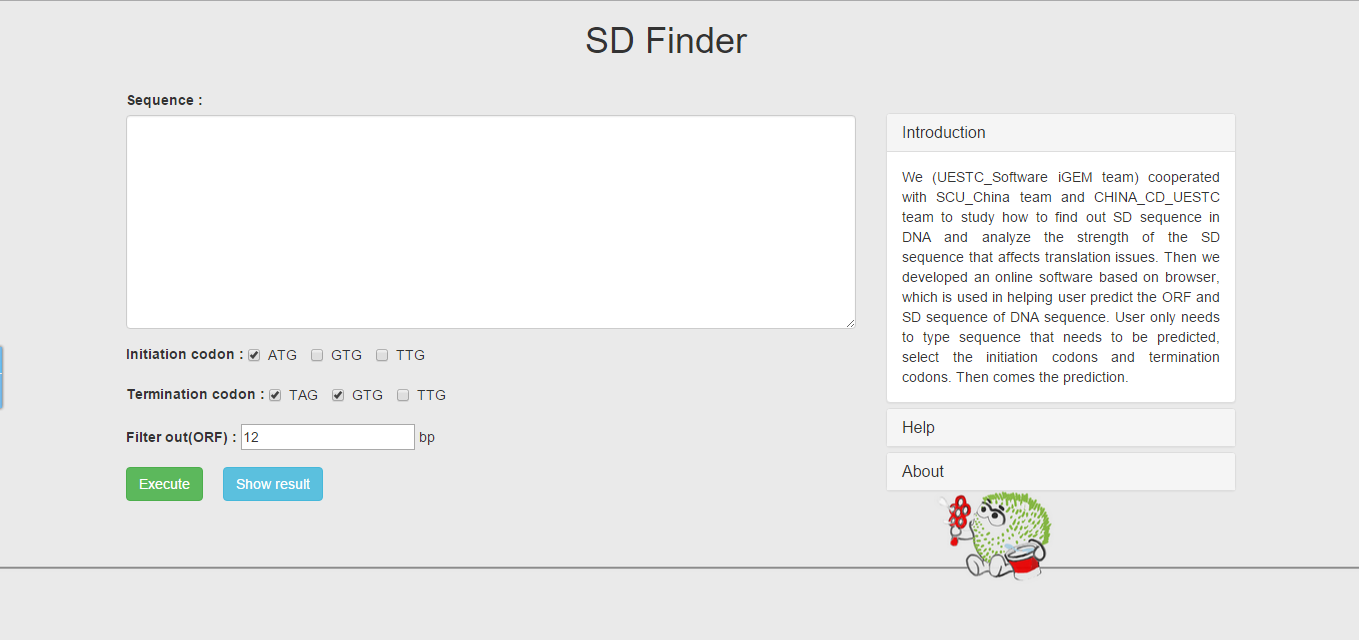
Online software based on browser—SD finder
We provide the related information about ORF and SD sequence in the result page. The column of SD sequence lists a 13bp sequence from -2 to -14 in upstream of ORF. We also tag the position of SD sequence signals with red. And the last column lists the type of the strength of the SD sequence.
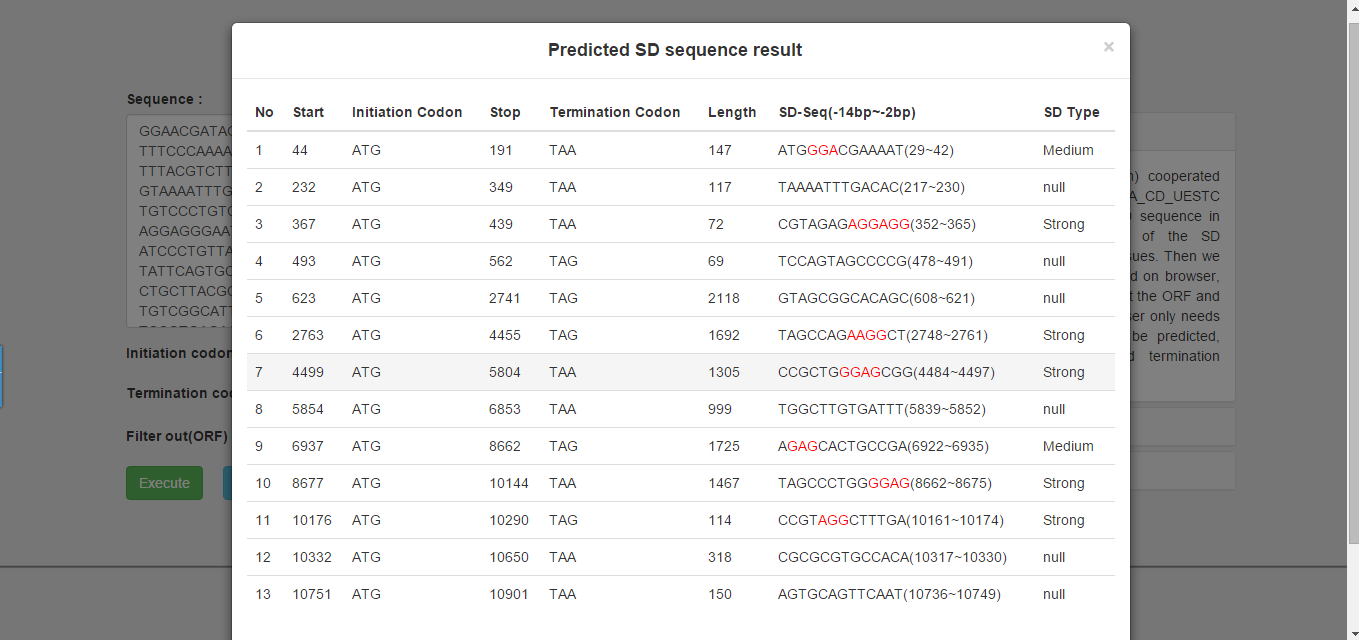
Result page
SD Finder: http://cefg.cn/Igem2015/mccap-server/app/tools/sdFinder.php
