|
|
| Line 1: |
Line 1: |
| | {{KU_Leuven}} | | {{KU_Leuven}} |
| | {{KU_Leuven/css}} | | {{KU_Leuven/css}} |
| − | {{KU_Leuven/symposium/test/css}} | + | {{KU_Leuven/wetlab/css}} |
| − | {{KU_Leuven/research/idea/css}} | + | {{KU_Leuven/main/css}} |
| − | {{KU_Leuven/Lightbox/css}}
| + | <html> |
| − | {{KU_Leuven/EasySwitch/CSS}}
| + | <head> |
| | | | |
| − |
| |
| − | <html>
| |
| | <script> | | <script> |
| | $(document).onload(function() { | | $(document).onload(function() { |
| Line 16: |
Line 14: |
| | <style> | | <style> |
| | | | |
| − | #interlab{ | + | #research{ |
| | background-color: transparent; | | background-color: transparent; |
| | border-style: solid; | | border-style: solid; |
| Line 23: |
Line 21: |
| | } | | } |
| | | | |
| − | #centernav:hover #interlab { /* this is active when your mouse moves is over the item */ | + | #centernav:hover #research { /* this is active when your mouse moves is over the item */ |
| | border: 0px solid transparent; | | border: 0px solid transparent; |
| | border-bottom: 0px solid transparent; | | border-bottom: 0px solid transparent; |
| | } | | } |
| | | | |
| − | .main-navm:hover #interlab { /* this is active when your mouse moves is over the item */ | + | .main-navm:hover #research { /* this is active when your mouse moves is over the item */ |
| | border: 0px solid transparent; | | border: 0px solid transparent; |
| | border-right: 0px solid transparent; | | border-right: 0px solid transparent; |
| Line 34: |
Line 32: |
| | | | |
| | @media screen and (max-width: 1000px) { | | @media screen and (max-width: 1000px) { |
| − | #interlab { | + | #research { |
| | border-bottom: 5px solid transparent; | | border-bottom: 5px solid transparent; |
| | border-right: 5px solid #8b7a57; | | border-right: 5px solid #8b7a57; |
| Line 46: |
Line 44: |
| | </style> | | </style> |
| | | | |
| − | <style> | + | <link rel="icon" href="https://static.igem.org/mediawiki/2015/9/9c/Ku_Leuven_Favicon.gif" /> |
| − | h3{
| + | <link rel="shortcut icon" href="https://static.igem.org/mediawiki/2015/9/9c/Ku_Leuven_Favicon.gif" / |
| − | font-size: 2em;
| + | </head> |
| − | }
| + | <body> |
| − | h2{
| + | |
| − | font-size:2.3em;
| + | |
| − | line-height: 120%;
| + | |
| − | text-allign="center";
| + | |
| − | text-transform: none;
| + | |
| − | }
| + | |
| − | b1{
| + | |
| − | font-size:0.85em;
| + | |
| − | }
| + | |
| − | p2{
| + | |
| − | allign: "center";
| + | |
| − | margin-top: 10px;
| + | |
| − | margin-bottom: 0px;
| + | |
| − | margin-left: 10px;
| + | |
| − | margin-right: 10px;
| + | |
| − | text-allign="left";
| + | |
| − | }
| + | |
| | | | |
| − | #tab1 { display:inline-block;
| |
| − | margin-left: 40px; }
| |
| − | #tab2 { display:inline-block;
| |
| − | margin-left: 44px; }
| |
| − | #tab3 { display:inline-block;
| |
| − | margin-left: 47px; }
| |
| | | | |
| − | #interlabstudy{
| |
| − | background-color: transparent;
| |
| − | border-style: solid;
| |
| − | border: 0px solid transparent;
| |
| − | border-bottom: 5px solid #8b7a57;
| |
| − | }
| |
| | | | |
| − | #centernav:hover #interlabstudy{ /* this is active when your mouse moves is over the item */
| + | <div id="headimg"> |
| − | border-style: solid;
| + | <div id="topimg"> |
| − | border-bottom: 0px solid transparent;
| + | </div> |
| − | }
| + | |
| | | | |
| − | #content {
| + | <div id="hiddenimg"> |
| − | background-color:transparent;
| + | <img src="https://static.igem.org/mediawiki/2015/b/bb/KU_Leuven_Zebra2.PNG" width="100%"> |
| − | }
| + | </div> |
| | | | |
| − | .li{
| |
| − | text-allign:left;
| |
| − | allign:left;
| |
| − | }
| |
| | | | |
| − | .tg {border-collapse:collapse;border-spacing:0;border-color:#aaa; margin: 0px auto 40px auto; }
| + | </div> |
| − | .tg td{font-family:Arial, sans-serif;font-size:14px;padding:10px 5px;border-style:solid;border-width:1px;overflow:hidden;word-break:normal;border-color:#aaa;color:#333;background-color:#fff;}
| + | |
| − | .tg th{font-family:Arial, sans-serif;font-size:14px;font-weight:normal;padding:10px 5px;border-style:solid;border-width:1px;overflow:hidden;word-break:normal;border-color:#aaa;color:#fff;background-color:#f38630;} <!- change colour
| + | |
| − | .tg .tg-s6z2{text-align:center}
| + | |
| − | </style>
| + | |
| − |
| + | |
| − | <link rel="stylesheet" type="text/css"
| + | |
| − | href="https://2015.igem.org/Template:KU_Leuven/Lightbox/CSS?action=raw&ctype=text/css" />
| + | |
| − | </head>
| + | |
| − | <script type="text/javascript" src="http://yourjavascript.com/11518521655/lightbox.js"></script>
| + | |
| | | | |
| − | <body> | + | |
| − | <div class="summaryheader"> | + | <div id="summarywrapper"> |
| − | <div class="summaryimg">
| + | <div id="logowrapper"> |
| − | <img src="https://static.igem.org/mediawiki/2015/b/b7/KU_Leuven_Researchbanner.jpg" width="100%">
| + | <div id="logoiGEMKUL"> |
| − | <div class="head">
| + | <img src="https://static.igem.org/mediawiki/2015/2/23/Team_KU_Leuven_banner.jpg" alt="Logo" width="100%" > |
| − | <h2> Interlab study </h2>
| + | |
| − | </div>
| + | |
| − | </div>
| + | |
| | </div> | | </div> |
| | | | |
| | | | |
| − | <!------------------------------------------------------Begin Content---------------------------------------------------------> | + | <div id="center"> |
| − | <div class="summarytext1"> | + | <div id="pagetitle"> |
| − | <div class="part"> | + | <h2>Research</h2> |
| | + | </div> |
| | + | <div id="scrolldown" class="center"> |
| | + | <img src="https://static.igem.org/mediawiki/2015/1/1d/KU_Leuven_Arrows_wiki.png" alt="Scroll down" width="100%"> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | <h2>Results</h2>
| + | <div id="summarylogos"> |
| − | <p>
| + | |
| − | Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nunc nibh ligula, faucibus a elit id, egestas posuere est. Donec cursus enim non lacus dignissim, id vulputate leo tincidunt. Aliquam pretium luctus augue, nec tempor odio sagittis eu. Morbi rutrum suscipit purus a iaculis. Suspendisse in augue eget turpis volutpat bibendum. Vivamus vel libero ac velit pulvinar auctor in eu nunc. Duis urna lectus, cursus ac tempor sed, lobortis id tellus.
| + | |
| − | </p>
| + | |
| − | <br>
| + | |
| − | <p>
| + | |
| − | <b>Table 1</b>
| + | |
| − | Fluorescence measurements
| + | |
| − | </p>
| + | |
| − | </br>
| + | |
| − | <div class="tg-wrap" style="width:100%"><table class="tg" style="width:100%"> | + | |
| − | <tr>
| + | |
| − | <th class="tg-031e">LB + Cam</th>
| + | |
| − | <th class="tg-031e">LB+Cam+Cells</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D1</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D2</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D3</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">512</td>
| + | |
| − | <td class="tg-031e">601</td>
| + | |
| − | <td class="tg-031e">737</td>
| + | |
| − | <td class="tg-031e">685</td>
| + | |
| − | <td class="tg-031e">751</td>
| + | |
| − | <td class="tg-031e">9425</td>
| + | |
| − | <td class="tg-031e">9322</td>
| + | |
| − | <td class="tg-031e">9737</td>
| + | |
| − | <td class="tg-031e">22786</td>
| + | |
| − | <td class="tg-031e">25895</td>
| + | |
| − | <td class="tg-031e">25048</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">546</td>
| + | |
| − | <td class="tg-031e">594</td>
| + | |
| − | <td class="tg-031e">754</td>
| + | |
| − | <td class="tg-031e">641</td>
| + | |
| − | <td class="tg-031e">641</td>
| + | |
| − | <td class="tg-031e">9669</td>
| + | |
| − | <td class="tg-031e">9545</td>
| + | |
| − | <td class="tg-031e">9876</td>
| + | |
| − | <td class="tg-031e">23159</td>
| + | |
| − | <td class="tg-031e">26460</td>
| + | |
| − | <td class="tg-031e">24785</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">549</td>
| + | |
| − | <td class="tg-031e">587</td>
| + | |
| − | <td class="tg-031e">722</td>
| + | |
| − | <td class="tg-031e">660</td>
| + | |
| − | <td class="tg-031e">697</td>
| + | |
| − | <td class="tg-031e">9407</td>
| + | |
| − | <td class="tg-031e">9321</td>
| + | |
| − | <td class="tg-031e">9677</td>
| + | |
| − | <td class="tg-031e">22719</td>
| + | |
| − | <td class="tg-031e">25931</td>
| + | |
| − | <td class="tg-031e">24935</td>
| + | |
| − | </tr>
| + | |
| − | <p>D1, D2, D3 <span id="tab1">Devices J23117 + I13504, J2310 + I13504 and J23101 + I13504 respectively</span></p>
| + | |
| − | <br>
| + | |
| − | </table></div>
| + | |
| | | | |
| − | <p> | + | <div id="summarylogotwitter"> |
| − | Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nunc nibh ligula, faucibus a elit id, egestas posuere est. Donec cursus enim non lacus dignissim, id vulputate leo tincidunt. Aliquam pretium luctus augue, nec tempor odio sagittis eu. Morbi rutrum suscipit purus a iaculis. Suspendisse in augue eget turpis volutpat bibendum. Vivamus vel libero ac velit pulvinar auctor in eu nunc. Duis urna lectus, cursus ac tempor sed, lobortis id tellus.
| + | <a href="https://twitter.com/kuleuven_igem" height="100%"><img src="https://static.igem.org/mediawiki/2015/b/b8/KU_Leuven_Twitter-logo.png" alt="Twitter" width="100%"></a> |
| − | </p> | + | </div> |
| − | <br> | + | |
| | | | |
| | + | <div id="summarylogofacebook"> |
| | + | <a href="https://www.facebook.com/KULeuveniGEM?fref=ts" alt="Facebook" height="50px"><img src="https://static.igem.org/mediawiki/2015/d/df/KU_Leuven_Fblogo.png" alt="mail" width="100%"></a> |
| | + | </div> |
| | + | |
| | + | <div id="summarylogomail"> |
| | + | <a href="mailto:igem@chem.kuleuven.be"><img src="https://static.igem.org/mediawiki/2015/1/11/KU_Leuven_mail.png" alt="mail" width="100%"></a> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="summary"> |
| | <p> | | <p> |
| − | <b>Table 2 </b> Absorbance measurements
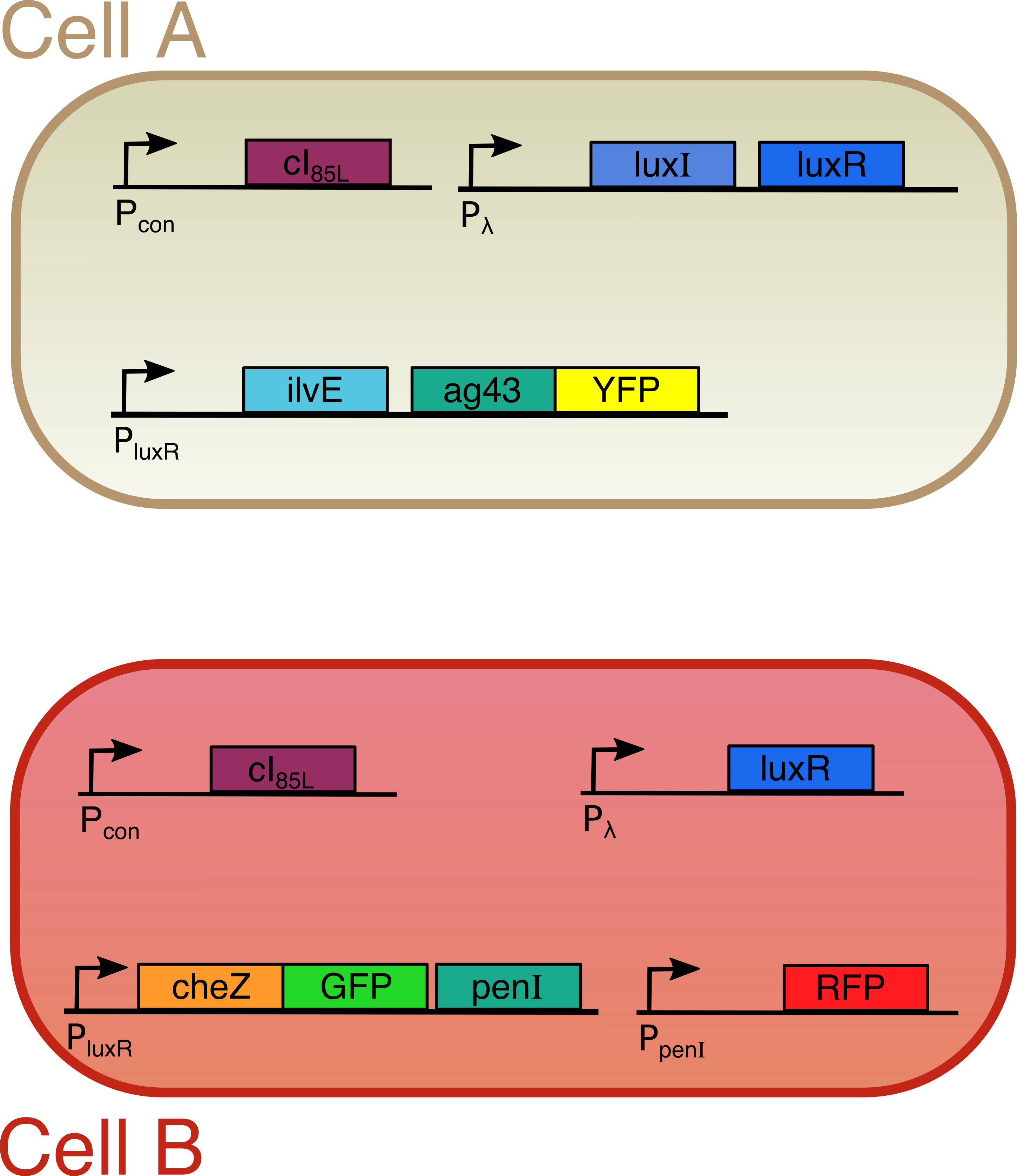
| + | We designed a circuit capable of forming patterns in a controlled way. Using a modified and temperature-sensitive lambda repressor (cI), we can trigger formation at desired points in time. This time-dependent controllability, together with the possibility to change many different parameters and output signals, leads to an enormous tunability in the creation of the patterns. Our mechanism will stimulate advancements in a variety of industrial processes like the creation of novel bio-materials. This fundamental project could also speed up medical research projects like tumor formation and tissue regeneration. |
| | </p> | | </p> |
| − | </br>
| + | <br/> |
| | + | <br/> |
| | + | <br/> |
| | + | <br/> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | <div class="tg-wrap" style="width:100%"><table class="tg" style="width:100%" > | + | <div class="subsections"> |
| − | <tr>
| + | <div class="subsectionwrapper"> |
| − | <th class="tg-031e">LB + Cam</th>
| + | <div class="subimgrow"> |
| − | <th class="tg-031e">LB+Cam+Cells</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D1</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D2</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D3</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">0.0434</td>
| + | |
| − | <td class="tg-031e">0.2052</td>
| + | |
| − | <td class="tg-031e">0.2174</td>
| + | |
| − | <td class="tg-031e">0.1942</td>
| + | |
| − | <td class="tg-031e">0.2201</td>
| + | |
| − | <td class="tg-031e">0.2088</td>
| + | |
| − | <td class="tg-031e">0.2082</td>
| + | |
| − | <td class="tg-031e">0.2154</td>
| + | |
| − | <td class="tg-031e">0.2139</td>
| + | |
| − | <td class="tg-031e">0.2210</td>
| + | |
| − | <td class="tg-031e">0.2218</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">0.0405</td>
| + | |
| − | <td class="tg-031e">0.2000</td>
| + | |
| − | <td class="tg-031e">0.2182</td>
| + | |
| − | <td class="tg-031e">0.1974</td>
| + | |
| − | <td class="tg-031e">0.1980</td>
| + | |
| − | <td class="tg-031e">0.2108</td>
| + | |
| − | <td class="tg-031e">0.2111</td>
| + | |
| − | <td class="tg-031e">0.2123</td>
| + | |
| − | <td class="tg-031e">0.2143</td>
| + | |
| − | <td class="tg-031e">0.2179</td>
| + | |
| − | <td class="tg-031e">0.2056</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">0.0379</td>
| + | |
| − | <td class="tg-031e">0.2064</td>
| + | |
| − | <td class="tg-031e">0.2141</td>
| + | |
| − | <td class="tg-031e">0.1961</td>
| + | |
| − | <td class="tg-031e">0.2036</td>
| + | |
| − | <td class="tg-031e">0.2137</td>
| + | |
| − | <td class="tg-031e">0.2117</td>
| + | |
| − | <td class="tg-031e">0.2164</td>
| + | |
| − | <td class="tg-031e">0.2202</td>
| + | |
| − | <td class="tg-031e">0.2261</td>
| + | |
| − | <td class="tg-031e">0.2083</td>
| + | |
| − | </tr>
| + | |
| − | <p>D1, D2, D3 <span id="tab1">Devices J23117 + I13504, J2310 + I13504 and J23101 + I13504 respectively</span></p>
| + | |
| − | <br>
| + | |
| − | </table>
| + | |
| | | | |
| − | <p> | + | <div class="subimg"> |
| − | Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nunc nibh ligula, faucibus a elit id, egestas posuere est. Donec cursus enim non lacus dignissim, id vulputate leo tincidunt. Aliquam pretium luctus augue, nec tempor odio sagittis eu. Morbi rutrum suscipit purus a iaculis. Suspendisse in augue eget turpis volutpat bibendum. Vivamus vel libero ac velit pulvinar auctor in eu nunc. Duis urna lectus, cursus ac tempor sed, lobortis id tellus.
| + | <a href="https://2015.igem.org/Team:KU_Leuven/Research/Idea"> |
| | + | <img src="https://static.igem.org/mediawiki/2015/4/4d/KU_Leuven_scheme_pattern_formation.png" width="95%" > |
| | + | </a> |
| | + | </div> |
| | + | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | + | |
| | + | <div class="subimg"> |
| | + | <img src="https://static.igem.org/mediawiki/2015/4/4e/KU_Leuven_Gblocks.jpg" width="100%"> |
| | + | </div> |
| | + | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | + | |
| | + | <div class="subimg"> |
| | + | <img src="https://static.igem.org/mediawiki/2015/3/32/KU_Leuven_AgarPlates.jpg" width="100%"> |
| | + | </div> |
| | + | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | + | |
| | + | <div class="subimg"> |
| | + | <img src="https://static.igem.org/mediawiki/2015/b/b7/KU_Leuven_Motility2.jpg" width="92.4%"> |
| | + | </div> |
| | + | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | + | |
| | + | </div> |
| | + | |
| | + | <div class="subtextrow"> |
| | + | <div class="subtext"> |
| | + | <a href="https://2015.igem.org/Team:KU_Leuven/Research/Idea"> |
| | + | <h2>Idea</h2> |
| | + | <p> A detailed description about the interaction between our two cells and the genetic circuit can be found here. |
| | + | <br/> |
| | + | <div class="more"><p>Read more</p></div> |
| | </p> | | </p> |
| − | <br> | + | </a> |
| | + | </div> |
| | | | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | + | |
| | + | <div class="subtext"> |
| | + | <h2>Parts</h2> |
| | <p> | | <p> |
| − | <b>Table 3</b> Values for Biological replicates
| + | More coming soon |
| | </p> | | </p> |
| − | </br> | + | <!---------<p>A list of all used biobricks, the modifications we performed on them and the new ones we designed. |
| − | <div class="tg-wrap" style="width:100%"><table class="tg" style="width:100%">
| + | <br/> |
| − | <tr>
| + | <a href="https://2015.igem.org/Team:KU_Leuven/Project/About">Read more</a> |
| − | <th class="tg-031e"></th>
| + | </p>-------> |
| − | <th class="tg-s6z2" colspan="3">D1</th>
| + | </div> |
| − | <th class="tg-s6z2" colspan="3">D2</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D3</th>
| + | <div class="whitespace"> |
| − | <th class="tg-s6z2">LB+Cam+Cells</th>
| + | </div> |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e"></td>
| + | |
| − | <td class="tg-031e">T1</td>
| + | |
| − | <td class="tg-031e">T2</td>
| + | |
| − | <td class="tg-031e">T3</td>
| + | |
| − | <td class="tg-031e">T1</td>
| + | |
| − | <td class="tg-031e">T2</td>
| + | |
| − | <td class="tg-031e">T3</td>
| + | |
| − | <td class="tg-031e">T1</td>
| + | |
| − | <td class="tg-031e">T2</td>
| + | |
| − | <td class="tg-031e">T2</td>
| + | |
| − | <td class="tg-031e"></td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">B1</td>
| + | |
| − | <td class="tg-031e">3390</td>
| + | |
| − | <td class="tg-031e">3456</td>
| + | |
| − | <td class="tg-031e">3372</td>
| + | |
| − | <td class="tg-031e">45139</td>
| + | |
| − | <td class="tg-031e">45868</td>
| + | |
| − | <td class="tg-031e">44020</td>
| + | |
| − | <td class="tg-031e">106526</td>
| + | |
| − | <td class="tg-031e">108068</td>
| + | |
| − | <td class="tg-031e">103174</td>
| + | |
| − | <td class="tg-031e">2929</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">B2</td>
| + | |
| − | <td class="tg-031e">3527</td>
| + | |
| − | <td class="tg-031e">3247</td>
| + | |
| − | <td class="tg-031e">3366</td>
| + | |
| − | <td class="tg-031e">44774</td>
| + | |
| − | <td class="tg-031e">45216</td>
| + | |
| − | <td class="tg-031e">44029</td>
| + | |
| − | <td class="tg-031e">117172</td>
| + | |
| − | <td class="tg-031e">121432</td>
| + | |
| − | <td class="tg-031e">114688</td>
| + | |
| − | <td class="tg-031e">2970</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">B3</td>
| + | |
| − | <td class="tg-031e">3399</td>
| + | |
| − | <td class="tg-031e">3237</td>
| + | |
| − | <td class="tg-031e">3426</td>
| + | |
| − | <td class="tg-031e">45204</td>
| + | |
| − | <td class="tg-031e">46519</td>
| + | |
| − | <td class="tg-031e">44718</td>
| + | |
| − | <td class="tg-031e">112931</td>
| + | |
| − | <td class="tg-031e">120550</td>
| + | |
| − | <td class="tg-031e">119707</td>
| + | |
| − | <td class="tg-031e">2844</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">Brep Avg</td>
| + | |
| − | <td class="tg-031e">3439</td>
| + | |
| − | <td class="tg-031e">3313</td>
| + | |
| − | <td class="tg-031e">3388</td>
| + | |
| − | <td class="tg-031e">45039</td>
| + | |
| − | <td class="tg-031e">45868</td>
| + | |
| − | <td class="tg-031e">44256</td>
| + | |
| − | <td class="tg-031e">112210</td>
| + | |
| − | <td class="tg-031e">116683</td>
| + | |
| − | <td class="tg-031e">112523</td>
| + | |
| − | <td class="tg-031e" rowspan="2"></td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">BRep SD</td>
| + | |
| − | <td class="tg-031e">270</td>
| + | |
| − | <td class="tg-031e">101</td>
| + | |
| − | <td class="tg-031e">162</td>
| + | |
| − | <td class="tg-031e">189</td>
| + | |
| − | <td class="tg-031e">532</td>
| + | |
| − | <td class="tg-031e">327</td>
| + | |
| − | <td class="tg-031e">4376</td>
| + | |
| − | <td class="tg-031e">6102</td>
| + | |
| − | <td class="tg-031e">6921</td>
| + | |
| − | </tr>
| + | |
| − | <p>D1, D2, D3 <span id="tab1">Devices J23117 + I13504, J2310 + I13504 and J23101 + I13504 respectively</span></p> | + | |
| − | <p>B1, B2, B3 <span id="tab2"> Biological replicates of the respective devices</span></p>
| + | |
| − | <p>T1, T2, T3 <span id="tab3"> Technical replicates</span></p> | + | |
| − | </br>
| + | |
| − | </table>
| + | |
| − | <p> | + | |
| − | Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nunc nibh ligula, faucibus a elit id, egestas posuere est. Donec cursus enim non lacus dignissim, id vulputate leo tincidunt. Aliquam pretium luctus augue, nec tempor odio sagittis eu. Morbi rutrum suscipit purus a iaculis. Suspendisse in augue eget turpis volutpat bibendum. Vivamus vel libero ac velit pulvinar auctor in eu nunc. Duis urna lectus, cursus ac tempor sed, lobortis id tellus.
| + | |
| − | </p>
| + | |
| − | <br>
| + | |
| − | <p>
| + | |
| − | <b> Table 4 </b> Values for Technical replicates
| + | |
| − | </p>
| + | |
| − | </br>
| + | |
| − | <table class="tg" style="width:100%">
| + | |
| − | <tr>
| + | |
| − | <th class="tg-031e"></th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D1</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D2</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D3</th>
| + | |
| − | <th class="tg-031e">LB+Cam+ Cells</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e"></td>
| + | |
| − | <td class="tg-031e">B1</td>
| + | |
| − | <td class="tg-031e">B2</td>
| + | |
| − | <td class="tg-031e">B3</td>
| + | |
| − | <td class="tg-031e">B1</td>
| + | |
| − | <td class="tg-031e">B2</td>
| + | |
| − | <td class="tg-031e">B3</td>
| + | |
| − | <td class="tg-031e">B1</td>
| + | |
| − | <td class="tg-031e">B2</td>
| + | |
| − | <td class="tg-031e">B3</td>
| + | |
| − | <td class="tg-031e"></td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">T1</td>
| + | |
| − | <td class="tg-031e">3390</td>
| + | |
| − | <td class="tg-031e">3527</td>
| + | |
| − | <td class="tg-031e">3399</td>
| + | |
| − | <td class="tg-031e">45139</td>
| + | |
| − | <td class="tg-031e">44774</td>
| + | |
| − | <td class="tg-031e">45204</td>
| + | |
| − | <td class="tg-031e">106526</td>
| + | |
| − | <td class="tg-031e">117172</td>
| + | |
| − | <td class="tg-031e">112931</td>
| + | |
| − | <td class="tg-031e">2929</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">T2</td>
| + | |
| − | <td class="tg-031e">3456</td>
| + | |
| − | <td class="tg-031e">3247</td>
| + | |
| − | <td class="tg-031e">3237</td>
| + | |
| − | <td class="tg-031e">45868</td>
| + | |
| − | <td class="tg-031e">45216</td>
| + | |
| − | <td class="tg-031e">46519</td>
| + | |
| − | <td class="tg-031e">108068</td>
| + | |
| − | <td class="tg-031e">121432</td>
| + | |
| − | <td class="tg-031e">120550</td>
| + | |
| − | <td class="tg-031e">2970</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">T3</td>
| + | |
| − | <td class="tg-031e">3372</td>
| + | |
| − | <td class="tg-031e">3366</td>
| + | |
| − | <td class="tg-031e">3426</td>
| + | |
| − | <td class="tg-031e">44020</td>
| + | |
| − | <td class="tg-031e">44029</td>
| + | |
| − | <td class="tg-031e">44718</td>
| + | |
| − | <td class="tg-031e">103174</td>
| + | |
| − | <td class="tg-031e">114688</td>
| + | |
| − | <td class="tg-031e">119707</td>
| + | |
| − | <td class="tg-031e">2844</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">TRep Avg</td>
| + | |
| − | <td class="tg-031e">3406</td>
| + | |
| − | <td class="tg-031e">3380</td>
| + | |
| − | <td class="tg-031e">3354</td>
| + | |
| − | <td class="tg-031e">45009</td>
| + | |
| − | <td class="tg-031e">44673</td>
| + | |
| − | <td class="tg-031e">45480</td>
| + | |
| − | <td class="tg-031e">105923</td>
| + | |
| − | <td class="tg-031e">117764</td>
| + | |
| − | <td class="tg-031e">117729</td>
| + | |
| − | <td class="tg-031e">2914</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">TRep SD</td>
| + | |
| − | <td class="tg-031e">36</td>
| + | |
| − | <td class="tg-031e">115</td>
| + | |
| − | <td class="tg-031e">140</td>
| + | |
| − | <td class="tg-031e">760</td>
| + | |
| − | <td class="tg-031e">490</td>
| + | |
| − | <td class="tg-031e">761</td>
| + | |
| − | <td class="tg-031e">2043</td>
| + | |
| − | <td class="tg-031e">2785</td>
| + | |
| − | <td class="tg-031e">3410</td>
| + | |
| − | <td class="tg-031e">52</td>
| + | |
| − | </tr>
| + | |
| | | | |
| − | <p>D1, D2, D3 <span id="tab1">Devices J23117 + I13504, J2310 + I13504 and J23101 + I13504 respectively</span></p> | + | <div class="subtext"> |
| − | <p>B1, B2, B3 <span id="tab2"> Biological replicates of the respective devices</span></p>
| + | <h2>Methods</h2> |
| − | <p>T1, T2, T3 <span id="tab3"> Technical replicates</span></p>
| + | |
| − | </br>
| + | |
| − | </table>
| + | |
| − | <div>
| + | |
| | <p> | | <p> |
| − | Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nunc nibh ligula, faucibus a elit id, egestas posuere est. Donec cursus enim non lacus dignissim, id vulputate leo tincidunt. Aliquam pretium luctus augue, nec tempor odio sagittis eu. Morbi rutrum suscipit purus a iaculis. Suspendisse in augue eget turpis volutpat bibendum. Vivamus vel libero ac velit pulvinar auctor in eu nunc. Duis urna lectus, cursus ac tempor sed, lobortis id tellus.
| + | More coming soon |
| | </p> | | </p> |
| − | <br> | + | <!---<p>Here you can find the performed steps to create two different cell types and detailed quantification methods to determine interesting parameters |
| | + | <br/> |
| | + | <a href="https://2015.igem.org/Team:KU_Leuven/Project/About">Read more</a> |
| | + | </p>-----> |
| | + | </div> |
| | | | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | + | |
| | + | <div class="subtext"> |
| | + | <!------ |
| | + | <a href="https://2015.igem.org/Team:KU_Leuven/Project/About">----> |
| | + | <h2>Results</h2> |
| | <p> | | <p> |
| − | <b> Table 5 </b> Standard graph
| + | More coming soon |
| | </p> | | </p> |
| | + | <!--------<p>The results of our experiments will appear here. |
| | + | <br/> |
| | + | <div class="more"><p> |
| | + | Read more</p> |
| | + | </div> |
| | + | </p>---------> |
| | + | </a> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | </br> | + | <div class="subimgrowm"> |
| − | <table class="tg" style="width:100%">
| + | |
| − | <tr>
| + | |
| − | <th class="tg-s6z2" colspan="7">Fluorescein standard</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">concentration (ng/mL)</td>
| + | |
| − | <td class="tg-031e">F1</td>
| + | |
| − | <td class="tg-031e">F2</td>
| + | |
| − | <td class="tg-031e">F3</td>
| + | |
| − | <td class="tg-031e">average F</td>
| + | |
| − | <td class="tg-031e">average F'</td>
| + | |
| − | <td class="tg-031e">SD</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">0</td>
| + | |
| − | <td class="tg-031e">9</td>
| + | |
| − | <td class="tg-031e">13</td>
| + | |
| − | <td class="tg-031e">12</td>
| + | |
| − | <td class="tg-031e">11</td>
| + | |
| − | <td class="tg-031e">0</td>
| + | |
| − | <td class="tg-031e">1.699673171</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">10</td>
| + | |
| − | <td class="tg-031e">1140</td>
| + | |
| − | <td class="tg-031e">1168</td>
| + | |
| − | <td class="tg-031e">1151</td>
| + | |
| − | <td class="tg-031e">1153</td>
| + | |
| − | <td class="tg-031e">1142</td>
| + | |
| − | <td class="tg-031e">11.5181017</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">25</td>
| + | |
| − | <td class="tg-031e">2543</td>
| + | |
| − | <td class="tg-031e">2600</td>
| + | |
| − | <td class="tg-031e">2631</td>
| + | |
| − | <td class="tg-031e">2591</td>
| + | |
| − | <td class="tg-031e">2580</td>
| + | |
| − | <td class="tg-031e">36.4447832</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">50</td>
| + | |
| − | <td class="tg-031e">4822</td>
| + | |
| − | <td class="tg-031e">5097</td>
| + | |
| − | <td class="tg-031e">5154</td>
| + | |
| − | <td class="tg-031e">5024</td>
| + | |
| − | <td class="tg-031e">5013</td>
| + | |
| − | <td class="tg-031e">144.9513328</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">125</td>
| + | |
| − | <td class="tg-031e">12879</td>
| + | |
| − | <td class="tg-031e">12902</td>
| + | |
| − | <td class="tg-031e">12931</td>
| + | |
| − | <td class="tg-031e">12904</td>
| + | |
| − | <td class="tg-031e">12893</td>
| + | |
| − | <td class="tg-031e">21.27596453</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">250</td>
| + | |
| − | <td class="tg-031e">25494</td>
| + | |
| − | <td class="tg-031e">25761</td>
| + | |
| − | <td class="tg-031e">25765</td>
| + | |
| − | <td class="tg-031e">25673</td>
| + | |
| − | <td class="tg-031e">25662</td>
| + | |
| − | <td class="tg-031e">126.8183303</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">375</td>
| + | |
| − | <td class="tg-031e">37474</td>
| + | |
| − | <td class="tg-031e">37505</td>
| + | |
| − | <td class="tg-031e">37899</td>
| + | |
| − | <td class="tg-031e">37626</td>
| + | |
| − | <td class="tg-031e">37615</td>
| + | |
| − | <td class="tg-031e">193.4545597</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">500</td>
| + | |
| − | <td class="tg-031e">46698</td>
| + | |
| − | <td class="tg-031e">49051</td>
| + | |
| − | <td class="tg-031e">48798</td>
| + | |
| − | <td class="tg-031e">48182</td>
| + | |
| − | <td class="tg-031e">48171</td>
| + | |
| − | <td class="tg-031e">1054.652023</td>
| + | |
| − | </tr>
| + | |
| | | | |
| − | </table> | + | <div class="subimg"> |
| | + | <b>Idea</b> |
| | + | <a href="https://2015.igem.org/Team:KU_Leuven/Research/Idea"> |
| | + | <img src="https://static.igem.org/mediawiki/2015/4/4d/KU_Leuven_scheme_pattern_formation.png" width="95%" > |
| | + | </a> |
| | + | </div> |
| | | | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | | | |
| − | <div class="center"> | + | <div class="subtext"> |
| − | <div id="image1">
| + | <a href="https://2015.igem.org/Team:KU_Leuven/Research/Idea"> |
| − |
| + | <p> A detailed description about the interaction between our two cells and the genetic circuit can be found here. |
| − | <img src="https://static.igem.org/mediawiki/2015/c/c2/KU_Leuven_Standardcurve.png" style="width:100%">
| + | </p> |
| − | <h4> | + | </a> |
| − | <div id=figure1> <b>Figure 1</b> </div>
| + | |
| − | Standard curve for fluorescein </h4>
| + | |
| | </div> | | </div> |
| | + | |
| | </div> | | </div> |
| | + | <div class="subimgrowm"> |
| | | | |
| | + | |
| | + | <div class="subimg"> |
| | + | <b>Parts</b> |
| | + | <img src="https://static.igem.org/mediawiki/2015/4/4e/KU_Leuven_Gblocks.jpg" width="100%"> |
| | + | </div> |
| | + | |
| | + | <div class="whitespace"> |
| | + | </div> |
| | + | |
| | + | <div class="subtext"> |
| | <p> | | <p> |
| − | Lorem ipsum dolor sit amet, consectetur adipiscing elit. Nunc nibh ligula, faucibus a elit id, egestas posuere est. Donec cursus enim non lacus dignissim, id vulputate leo tincidunt. Aliquam pretium luctus augue, nec tempor odio sagittis eu. Morbi rutrum suscipit purus a iaculis. Suspendisse in augue eget turpis volutpat bibendum. Vivamus vel libero ac velit pulvinar auctor in eu nunc. Duis urna lectus, cursus ac tempor sed, lobortis id tellus.
| + | More coming soon |
| | </p> | | </p> |
| − | <br> | + | <!---------<p>A list of all used biobricks, the modifications we performed on them and the new ones we designed. |
| − | <p> | + | <a href="https://2015.igem.org/Team:KU_Leuven/Project/About">Read more</a> |
| − | <b> Table 6 </b> Extrapolation of GFP concentration using fluorescence measurements and fluorescein standard | + | </p>-------> |
| − | </p> | + | </div> |
| − | </br> | + | |
| | | | |
| − | <table class="tg" style="width:100%">
| + | </div> |
| − | <tr>
| + | <div class="subimgrowm"> |
| − | <th class="tg-031e"></th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D1</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D2</th>
| + | |
| − | <th class="tg-s6z2" colspan="3">D3</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">Brep Avg</td>
| + | |
| − | <td class="tg-031e">3439</td>
| + | |
| − | <td class="tg-031e">3313</td>
| + | |
| − | <td class="tg-031e">3388</td>
| + | |
| − | <td class="tg-031e">45039</td>
| + | |
| − | <td class="tg-031e">45868</td>
| + | |
| − | <td class="tg-031e">44256</td>
| + | |
| − | <td class="tg-031e">112210</td>
| + | |
| − | <td class="tg-031e">116683</td>
| + | |
| − | <td class="tg-031e">112523</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">(Brep Avg) - (LB+cam+cells avg)</td>
| + | |
| − | <td class="tg-031e">525</td>
| + | |
| − | <td class="tg-031e">399</td>
| + | |
| − | <td class="tg-031e">474</td>
| + | |
| − | <td class="tg-031e">42125</td>
| + | |
| − | <td class="tg-031e">42954</td>
| + | |
| − | <td class="tg-031e">41342</td>
| + | |
| − | <td class="tg-031e">109296</td>
| + | |
| − | <td class="tg-031e">113769</td>
| + | |
| − | <td class="tg-031e">109609</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">Concentration (ng/mL)</td>
| + | |
| − | <td class="tg-031e">1.929</td>
| + | |
| − | <td class="tg-031e">0.639</td>
| + | |
| − | <td class="tg-031e">1.407</td>
| + | |
| − | <td class="tg-031e">427.88</td>
| + | |
| − | <td class="tg-031e">436.36</td>
| + | |
| − | <td class="tg-031e">419.88</td>
| + | |
| − | <td class="tg-031e">1115.64</td>
| + | |
| − | <td class="tg-031e">1161.44</td>
| + | |
| − | <td class="tg-031e">1118.85</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">TRep Avg</td>
| + | |
| − | <td class="tg-031e">3406</td>
| + | |
| − | <td class="tg-031e">3380</td>
| + | |
| − | <td class="tg-031e">3354</td>
| + | |
| − | <td class="tg-031e">45009</td>
| + | |
| − | <td class="tg-031e">44673</td>
| + | |
| − | <td class="tg-031e">45480</td>
| + | |
| − | <td class="tg-031e">105923</td>
| + | |
| − | <td class="tg-031e">117764</td>
| + | |
| − | <td class="tg-031e">117729</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">(Trep avg) - (LB+cam+cellsavg)</td>
| + | |
| − | <td class="tg-031e">492</td>
| + | |
| − | <td class="tg-031e">466</td>
| + | |
| − | <td class="tg-031e">440</td>
| + | |
| − | <td class="tg-031e">42095</td>
| + | |
| − | <td class="tg-031e">41759</td>
| + | |
| − | <td class="tg-031e">42566</td>
| + | |
| − | <td class="tg-031e">103009</td>
| + | |
| − | <td class="tg-031e">114850</td>
| + | |
| − | <td class="tg-031e">114815</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-031e">Concentration (ng/mL)</td>
| + | |
| − | <td class="tg-031e">1.591</td>
| + | |
| − | <td class="tg-031e">1.325</td>
| + | |
| − | <td class="tg-031e">1.059</td>
| + | |
| − | <td class="tg-031e">427.56</td>
| + | |
| − | <td class="tg-031e">424.12</td>
| + | |
| − | <td class="tg-031e">432.39</td>
| + | |
| − | <td class="tg-031e">1051.27</td>
| + | |
| − | <td class="tg-031e">1172.51</td>
| + | |
| − | <td class="tg-031e">1172.15</td>
| + | |
| − | </tr>
| + | |
| − | </table>
| + | |
| | | | |
| | | | |
| | + | <div class="subimg"> |
| | + | <b>Methods</b> |
| | + | <img src="https://static.igem.org/mediawiki/2015/3/32/KU_Leuven_AgarPlates.jpg" width="100%"> |
| | </div> | | </div> |
| | + | <div class="whitespace"> |
| | </div> | | </div> |
| | | | |
| | + | <div class="subtext"> |
| | + | <p> |
| | + | More coming soon |
| | + | </p> |
| | + | <!---<p>Here you can find the performed steps to create two different cell types and detailed quantification methods to determine interesting parameters |
| | + | <a href="https://2015.igem.org/Team:KU_Leuven/Project/About">Read more</a> |
| | + | </p>-----> |
| | + | </div> |
| | | | |
| − | <div class="summarytext1"> | + | </div> |
| − | <div class="part"> | + | <div class="subimgrowm"> |
| − | <h2>Protocols</h2>
| + | |
| | | | |
| − | <p> | + | <div class="subimg"> |
| | + | <b>Results</b> |
| | + | <img src="https://static.igem.org/mediawiki/2015/b/b7/KU_Leuven_Motility2.jpg" width="92.4%"> |
| | + | </div> |
| | + | <div class="whitespace"> |
| | + | </div> |
| | | | |
| − | <br> | + | <div class="subtext"> |
| − | <br> | + | <!------ |
| − | <br> | + | <a href="https://2015.igem.org/Team:KU_Leuven/Project/About">----> |
| − | <br> | + | <p> |
| | + | More coming soon |
| | </p> | | </p> |
| | + | <!--------<p>The results of our experiments will appear here. |
| | + | <br/> |
| | + | <div class="more"><p> |
| | + | Read more</p> |
| | + | </div> |
| | + | </p>---------> |
| | + | </a> |
| | + | </div> |
| | </div> | | </div> |
| | </div> | | </div> |
| Line 665: |
Line 290: |
| | | | |
| | | | |
| − |
| |
| − | </br>
| |
| − |
| |
| − | <!--------------------------------------------------EasySwitch-------------------------------------------------------------->
| |
| − | <script language="javascript" type="text/javascript">
| |
| − | function EasySwitch(){
| |
| − | var lckv= document.getElementById("EasySwitch").checked;
| |
| − | if (lckv==true)
| |
| − | {
| |
| − | $('.easy').css('display', 'inline');
| |
| − | $('.scientific').css('display', 'none');
| |
| − | }
| |
| − | else
| |
| − | {
| |
| − | $('.easy').css('display', 'none');
| |
| − | $('.easyleft').css('display', 'none');
| |
| − | $('.scientific').css('display', 'inline');
| |
| − | }
| |
| − | }
| |
| − |
| |
| − | $( "#Switch" ).mouseenter(function() {
| |
| − | $("#SwitchText").show();
| |
| − | });
| |
| − | $( "#Switch" ).mouseleave(function() {
| |
| − | $("#SwitchText").hide();
| |
| − | });
| |
| − | </script>
| |
| | </body> | | </body> |
| | </html> | | </html> |
| | {{KU_Leuven/modeling/internal/ultra/test/footer}} | | {{KU_Leuven/modeling/internal/ultra/test/footer}} |