Difference between revisions of "Team:Chalmers-Gothenburg/Project"
| (17 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{ | + | {{Chalmers-Gothenburg}} |
| − | < | + | <h2> Project </h2> |
| − | < | + | |
| − | + | <p>Contaminations – a problem you definitely want to get rid of as quickly as possible. It can destroy your product and lower the efficiency of the process, which in the end may lead to higher expenses. We have created a great system for both finding and combating contaminations in bioreactors, automatically! Our intention was to deal with the hard process in finding unwanted cells in bioreactors and our solution is an innovative system that is both time and material efficient. With our project you no longer need to worry about long analytic times and if your product is pure enough, it will all be taken care of directly in the process! Do you know what the best part is? The system is really cool and can be used for so much more!</p> | |
| − | + | ||
| − | + | <p>Our project presents a solution for dealing with contamination in bioreactors. We have found a way to detect and deal with contamination without wasting product in the tank. The solution could be used both for academic and industrial use. We have designed a two system solution that we have implemented in <i>Saccharomyces cerevisiae</i>, a solution that is both efficient and time saving. The systems are based on one detection system that gives a fluorescent signal when a contaminant is present and one DNA-repair-system. The detection system uses pheromone detection to sense the present of a contaminant and uses an intracellular cascade reaction to send out a RFP-signal. The repair system is based on a system from the bacterium <i>Deinococcus radiodurans</i> and repairs blunt DNA-cuts, which are hard for the cell to repair. This system makes it possible to irradiate the tank with UV-irradiation that will kill the contaminant but not the yeast. These systems together with a safety switch for use in research will be presented in detail in this chapter.</p> <p>Figure 1-6 illustrates the general concept of our project.</p> | |
| − | + | [[File:ChalmersGothenburgPurTHEORY1.png|400px|]] | |
| − | + | <p class="figuretext"><b>Figure 1.</b> The bioreactor in its initial state containing only the producing organism(yellow), <i>Saccharomyces cerevisiae</i>.</p> | |
| − | + | ||
| − | + | ||
| − | + | [[File:ChalmersGothenburgPurTHEORY2.png|400px|]] | |
| − | + | <p class="figuretext"><b>Figure 2.</b> Contaminants(blue) getting into the reactor. </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | [[File:ChalmersGothenburgPurTHEORY3.png|400px|]] | |
| − | + | <p class="figuretext"><b>Figure 3.</b>Producing <i>S. cerevisiae</i> cells expressing RFP(red) upon detection, signaling the presence of contaminants. </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | [[File:ChalmersGothenburgPurTHEORY4.png|400px|]] | |
| − | + | <p class="figuretext"><b>Figure 4.</b> Detection signal initiating the irradiation process, its purpose being to combat the contaminants. </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | [[File:ChalmersGothenburgPurTHEORY5.png|400px|]] | |
| − | + | <p class="figuretext"><b>Figure 5.</b> Only modified <i>S. cerevisiae</i> cells survive the radiation due to the implemented repair system. Contaminants and modified cells that have expressed the detection signal are washed out. </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | [[File:ChalmersGothenburgPurTHEORY1.png|400px|]] | |
| + | <p class="figuretext"><b>Figure 6.</b> The bioreactor eventually returns to its original state.</p> | ||
| − | + | <html> | |
| − | + | </div> | |
| − | + | </div> | |
| − | + | </div> | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 01:03, 19 September 2015


Project
Contaminations – a problem you definitely want to get rid of as quickly as possible. It can destroy your product and lower the efficiency of the process, which in the end may lead to higher expenses. We have created a great system for both finding and combating contaminations in bioreactors, automatically! Our intention was to deal with the hard process in finding unwanted cells in bioreactors and our solution is an innovative system that is both time and material efficient. With our project you no longer need to worry about long analytic times and if your product is pure enough, it will all be taken care of directly in the process! Do you know what the best part is? The system is really cool and can be used for so much more!
Our project presents a solution for dealing with contamination in bioreactors. We have found a way to detect and deal with contamination without wasting product in the tank. The solution could be used both for academic and industrial use. We have designed a two system solution that we have implemented in Saccharomyces cerevisiae, a solution that is both efficient and time saving. The systems are based on one detection system that gives a fluorescent signal when a contaminant is present and one DNA-repair-system. The detection system uses pheromone detection to sense the present of a contaminant and uses an intracellular cascade reaction to send out a RFP-signal. The repair system is based on a system from the bacterium Deinococcus radiodurans and repairs blunt DNA-cuts, which are hard for the cell to repair. This system makes it possible to irradiate the tank with UV-irradiation that will kill the contaminant but not the yeast. These systems together with a safety switch for use in research will be presented in detail in this chapter.
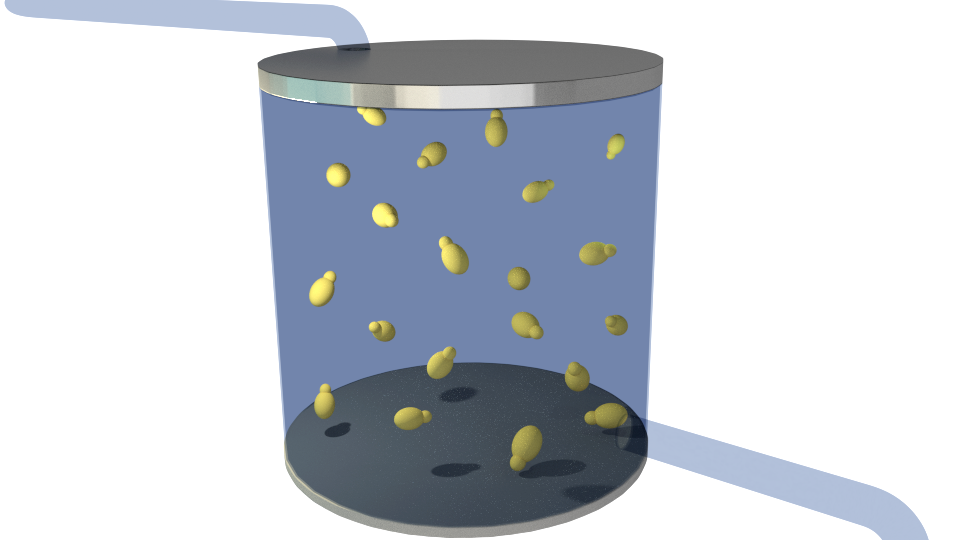
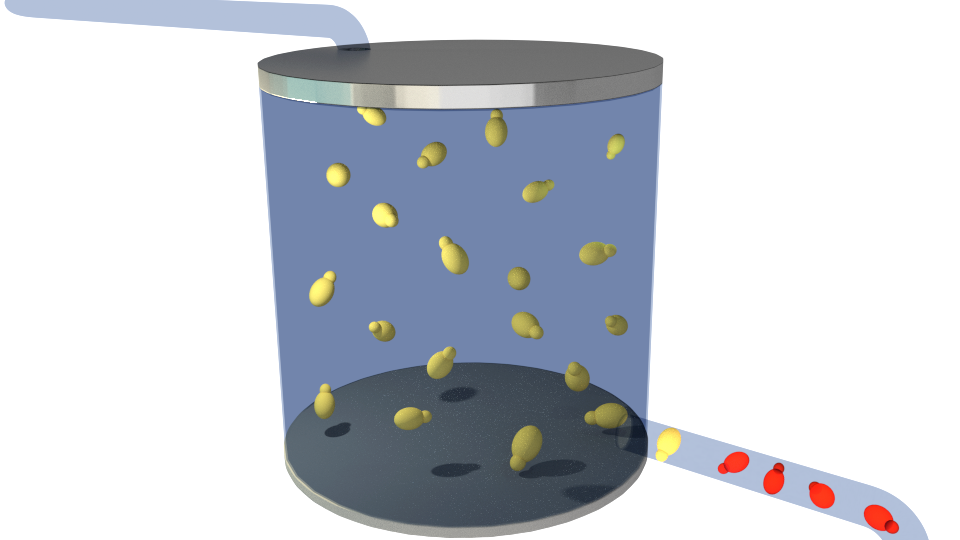
Figure 1-6 illustrates the general concept of our project.
Figure 1. The bioreactor in its initial state containing only the producing organism(yellow), Saccharomyces cerevisiae.
Figure 2. Contaminants(blue) getting into the reactor.
Figure 3.Producing S. cerevisiae cells expressing RFP(red) upon detection, signaling the presence of contaminants.
Figure 4. Detection signal initiating the irradiation process, its purpose being to combat the contaminants.
Figure 5. Only modified S. cerevisiae cells survive the radiation due to the implemented repair system. Contaminants and modified cells that have expressed the detection signal are washed out.
Figure 6. The bioreactor eventually returns to its original state.