Difference between revisions of "Team:Chalmers-Gothenburg/Project Results"
| Line 333: | Line 333: | ||
<p>Construct 1 was successfully assembled and transformed into E.coli, see figure 2, but the construct is not functional without construct 2 and 3. Because of this limitation, no further steps were taken with construct 1.</p> | <p>Construct 1 was successfully assembled and transformed into E.coli, see figure 2, but the construct is not functional without construct 2 and 3. Because of this limitation, no further steps were taken with construct 1.</p> | ||
[[File:ChalmersGothenburgDCas92.jpg]] | [[File:ChalmersGothenburgDCas92.jpg]] | ||
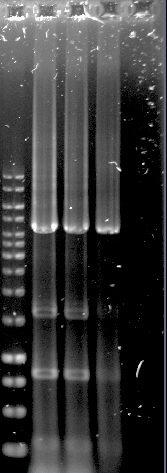
| − | <p> <b>Figure 1.Well 1: 1kb ladder. Well 2 and 3: unpurified dCas9-Vp64. Well 4: purified dCas9-Vp64. Expected length: 4335 bp</b> | + | <p> <b>Figure 1.Well 1: 1kb ladder. Well 2 and 3: unpurified dCas9-Vp64. Well 4: purified dCas9-Vp64. Expected length: 4335 bp.</b></p> |
<p>Construct 1 was successfully assembled and transformed into E.coli, see figure 2, but the construct is not functional without construct 2 and 3. Because of this limitation, no further steps were taken with construct 1.</p> | <p>Construct 1 was successfully assembled and transformed into E.coli, see figure 2, but the construct is not functional without construct 2 and 3. Because of this limitation, no further steps were taken with construct 1.</p> | ||
[[File:ChalmersGothenburgVerifyC1.jpeg]] | [[File:ChalmersGothenburgVerifyC1.jpeg]] | ||
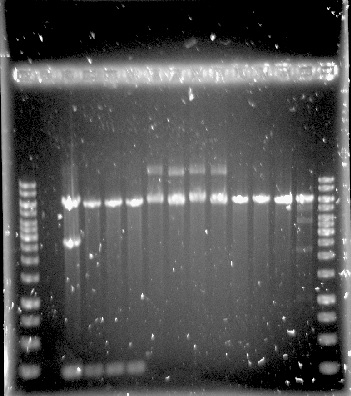
| − | <p><b>Figure 2. Construct 1 digested with MfeI. Expected fragments: 214, 2656 and 5926 bp</b></p> | + | <p><b>Figure 2. Construct 1 digested with MfeI. Expected fragments: 214, 2656 and 5926 bp.</b></p> |
<h3>Detection test</h3> | <h3>Detection test</h3> | ||
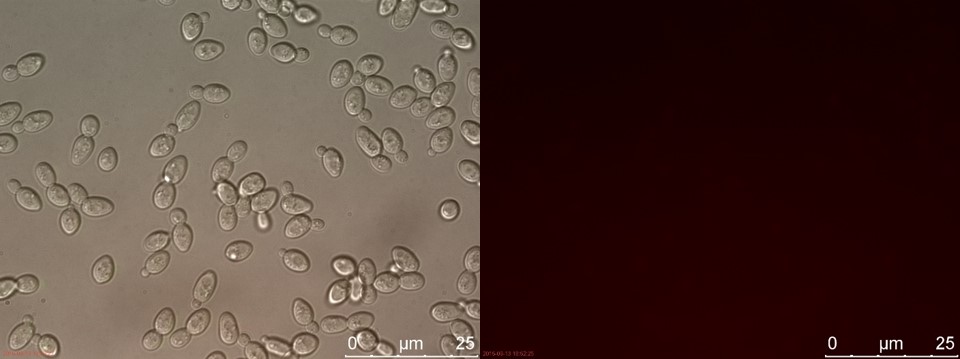
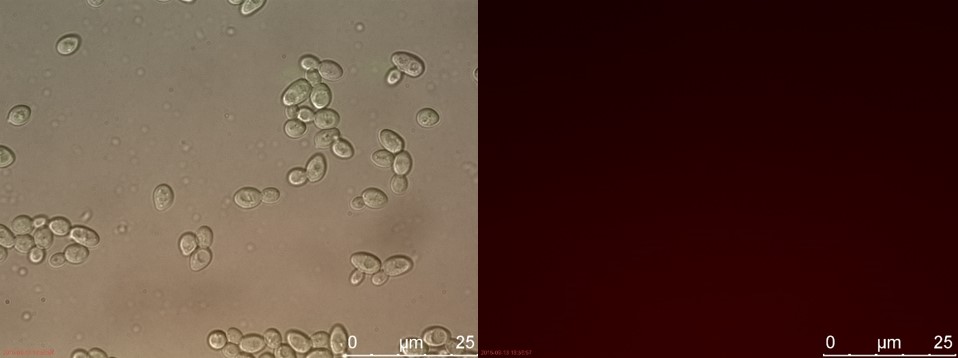
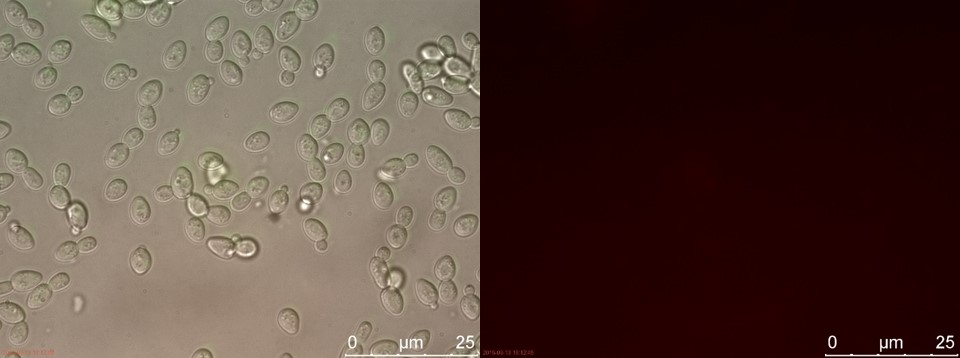
| − | <p>The fluorescent microscopy pictures from the detection test with CEN.PK2 containing construct 4 (C4) are shown in figure | + | <p>The fluorescent microscopy pictures from the detection test with CEN.PK2 containing construct 4 (C4) are shown in figure 3-6. Samples were made with different amounts of concentrated P-factor and compared with wild type CEN.PK2 (WT).<br>Only the C4 samples containing the highest amount of P-factor showed a few fluorescent cells, which is promising for our concept. The weak signal can be explained by the fact that constructs containing the amplification system through dCas9-vp64 could not be completed. This forces the detection system to rely on the weak promoter pFUS1 <b>[1]</b> to express mRFP. This can result in a weak fluorescent signal when the P-factor is detected.</p> |
| + | <p>[[File:ChalmersGothenburgC4230.jpg]] | ||
| + | <br><b>Figure 3. C4 with 230 µl concentrated P-factor</b></p> | ||
| + | <p>[[File:ChalmersGothenburgC40.jpg]] | ||
| + | <br><b>Figure 4. C4 without P-factor</b></p> | ||
| + | <p>[[File:ChalmersGothenburgWt230.jpg]] | ||
| + | <br><b>Figure 5. WT with 230 µl concentrated P-factor</b></p> | ||
| + | <p>[[File:ChalmersGothenburgWt0.jpg]] | ||
| + | <br><b>Figure 5. WT without P-factor</b></p> | ||
Revision as of 17:04, 18 September 2015
Overview
• Assembled and integrated two different safety switches, based on TPK2 overexpression or mRFP expression induced at low ATP levels, into CEN.PK2.
• SS TPK2 shows reduced growth rate after OD-measurement but no change in viability compared to wild type.
• mRFP measurements show that connection of pTEF1 to pSUC2 maintains the ATP repression mechanism of pSUC2, while achieving higher expression at low ATP levels.
• Successfully constructed and integrated the system to detect the P-factor from S.pombe into CEN.PK2.
• Fluorescent cells in DAS
• Amplified all parts of the PUR system, but obtained vector-only clones after Gibson and transformation into E.coli
DAS
The system without the amplification through dCAS9-VP64 (construct 4) was successfully assembled and integrated into S.cerevisiae CEN.PK2. The genomic integration was verified with colony PCR and sequencing. The detection system was initially intended to be integrated into IMFD-70, but as several transformations failed the strain was changed to CEN.PK2 instead.
The construction of the application systems (construct 2 and 3) was unsuccessful. The fragments of the constructs was individually amplified with PCR, see figure 1, but only clones with empty vectors were obtained after Gibson assembly and transformation into E.coli.
Construct 1 was successfully assembled and transformed into E.coli, see figure 2, but the construct is not functional without construct 2 and 3. Because of this limitation, no further steps were taken with construct 1.
Figure 1.Well 1: 1kb ladder. Well 2 and 3: unpurified dCas9-Vp64. Well 4: purified dCas9-Vp64. Expected length: 4335 bp.
Construct 1 was successfully assembled and transformed into E.coli, see figure 2, but the construct is not functional without construct 2 and 3. Because of this limitation, no further steps were taken with construct 1.
Figure 2. Construct 1 digested with MfeI. Expected fragments: 214, 2656 and 5926 bp.
Detection test
The fluorescent microscopy pictures from the detection test with CEN.PK2 containing construct 4 (C4) are shown in figure 3-6. Samples were made with different amounts of concentrated P-factor and compared with wild type CEN.PK2 (WT).
Only the C4 samples containing the highest amount of P-factor showed a few fluorescent cells, which is promising for our concept. The weak signal can be explained by the fact that constructs containing the amplification system through dCas9-vp64 could not be completed. This forces the detection system to rely on the weak promoter pFUS1 [1] to express mRFP. This can result in a weak fluorescent signal when the P-factor is detected.
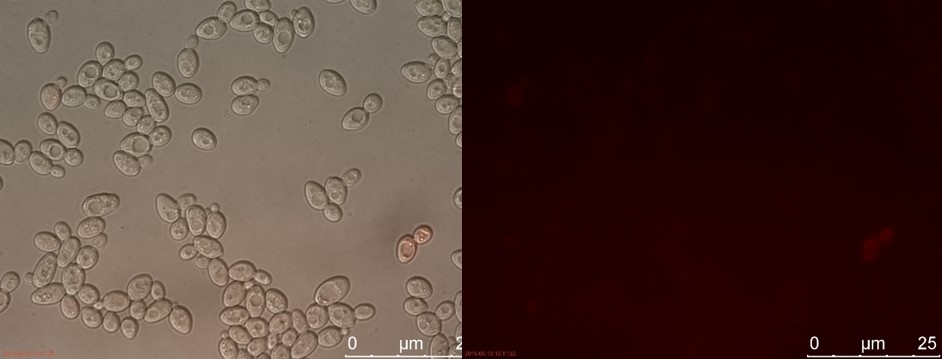
Figure 3. C4 with 230 µl concentrated P-factor