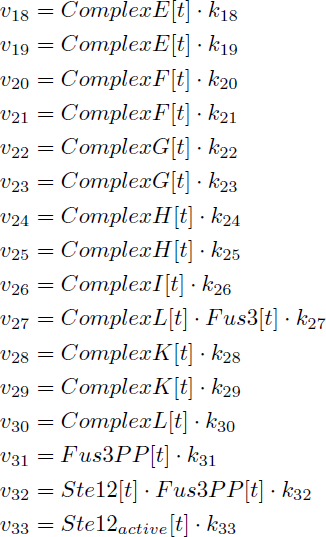Team:Chalmers-Gothenburg/Detection


Detection
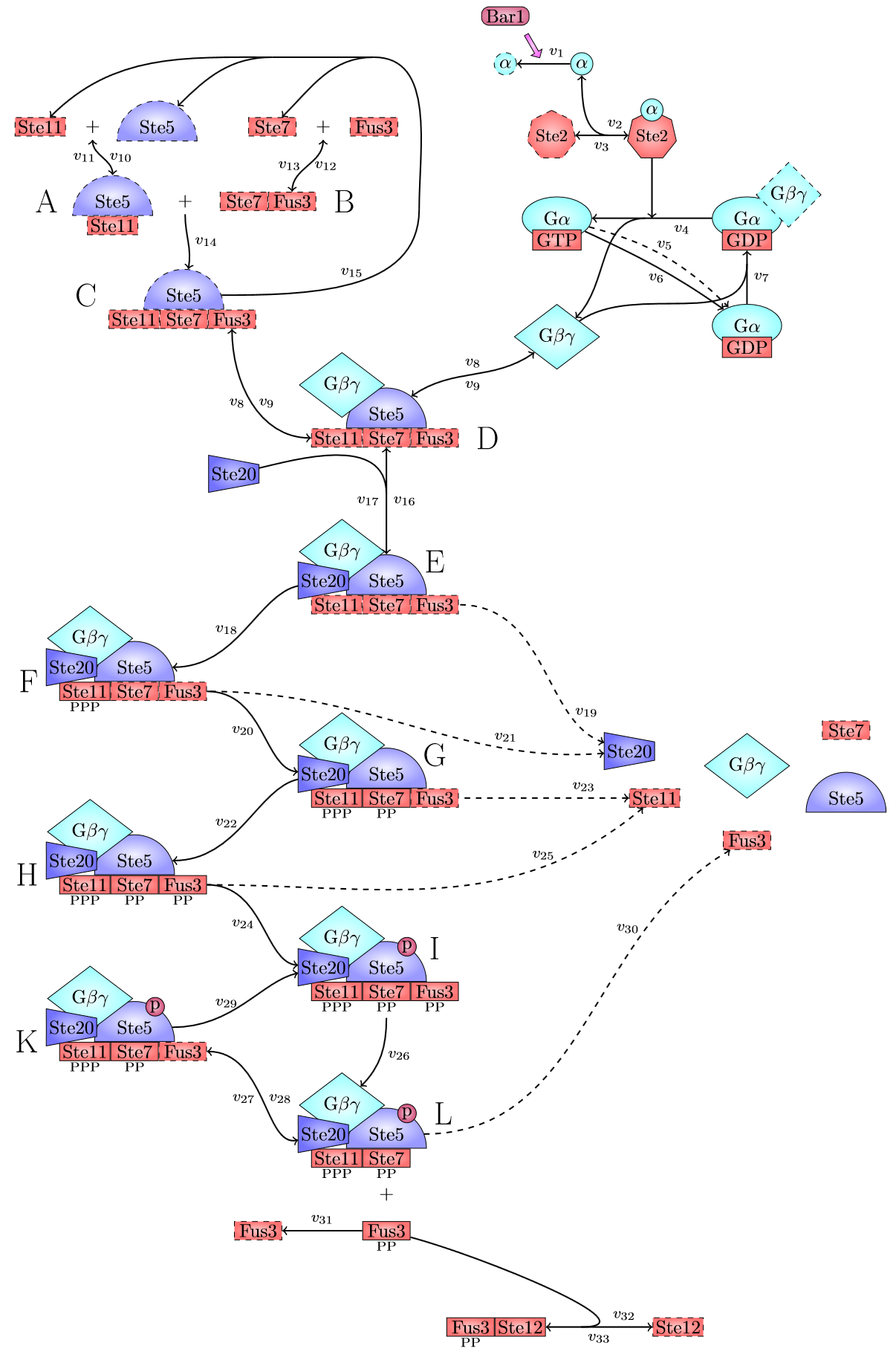
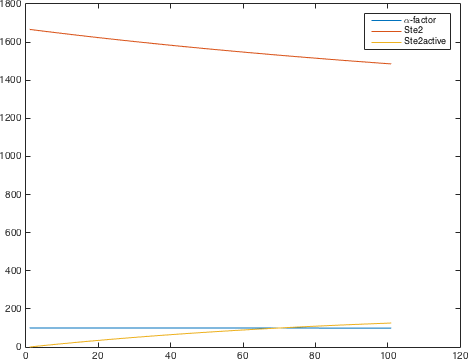
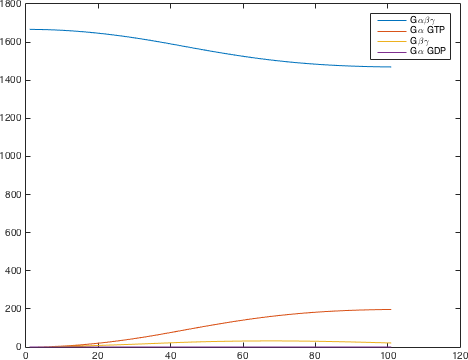
The detection system is used so allow Saccharomyces cerevisiae to detect a foreign peptide. The system couples a signaling pathway, the pheromone pathway with a receptor in from a species from a different another Subphylum. The pheromone pathway starts with a ligand receptor association followed by disassociation of g proteins associated to the receptor. The disassociated g- protein starts a phosphorylation cascade (MAPK) leading to a activation of a transcriptionfactor.
Model Formation
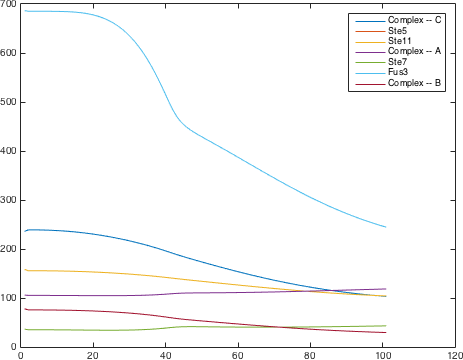
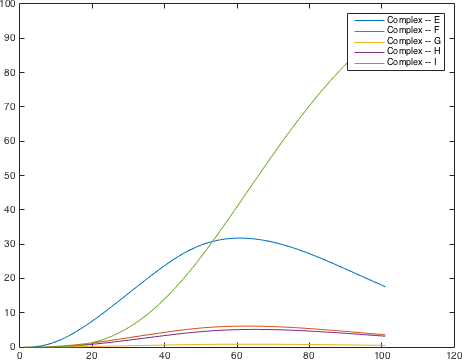
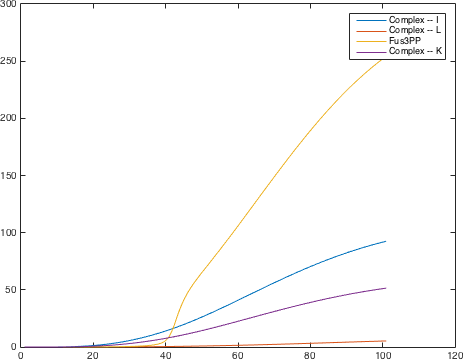
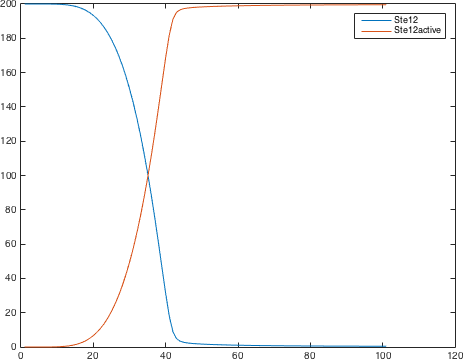
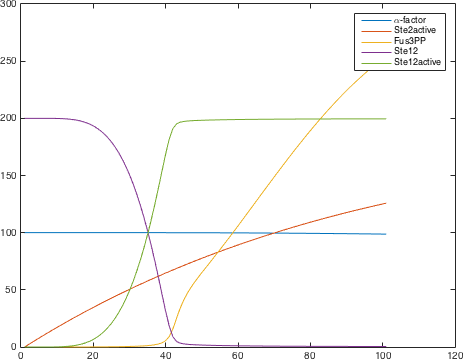
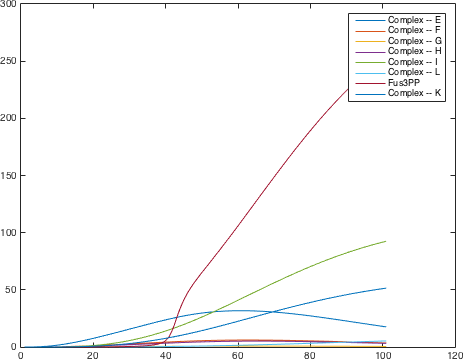
There are significant work on modeling the pheromone pathway, there is even a database dedicated to yeast pheromone pathway modeling (http://yeastpheromonemodel.org/). The receptor is a G - protein coupled receptor (GPCR), the G-proteins are the Gα, Gβ, and Gγ, the Gβ and Gγ proteins interactions are neglected in the model and the proteins are assumed to always be associated. A similar assumption is made on the Gα protein and its associated GDP, however, the GDP can still me phosphorylated to GTP. All the monomers present in the MAPK-cascade complexes were assumed not to be associated all the time. Also all MAPK-cascade were assumed to irreversible reactions except from the last reaction where Fus3 can re associate with the complex and then reenter the cascade. The activation Ste12 by the inhibition of Dig1 and Dig2 is a one-step reaction between Fus3pp and inactive Ste12. The Protein expression were assumed to be linear to the amount of active transcription factor.
The reactions was defines as:
Yielding the following differential equations:
Parameters
| [Ste5] | 158.33 nM |
| [Ste11] | 158.33 nM |
| [Ste7] | 36.40 nM |
| [Fus3] | 686.40 nM |
| [Ste12] | 200 nM |
| [Ste20] | 1000 nM |
| [Gαβγ] | 1666.67 nM |
| [Ste5/Ste11] | 105.94 nM |
| [Ste7/Fus3] | 77.87 nM |
| [Ste5/Ste11/Ste7/Fus3] | 235.72 nM |
| Rest | 0 |
| k1 | 0.03min−1 nM−1 | [1],[2] |
| k2 | 0.0012 min−1 nM−1 | [1],[2] |
| k3 | 0.6 min−1 | [1],[2] |
| k4 | 0.24 min−1 | [1],[2] |
| k5 | 0.024 min−1 | [1],[2] |
| k6 | 0.0036 min−1 nM−1 | [1],[2] |
| k7 | 0.24 min−1 | [1],[2] |
| k8 | 0.33min−1 nM−1 | [1],[2] |
| k9 | 2000 min−1 nM−1 | [1],[2] |
| k10 | 0.1 min−1 nM−1 | [1] |
| k11 | 5min−1 | [1] |
| k12 | 1min−1 nM−1 | [1] |
| k13 | 3min−1 | [1] |
| k14 | 1min−1 nM−1 | [1],[3] |
| k15 | 3min−1 | [1],[3] |
| k16 | 3min−1 nM−1 | [1] |
| k17 | 100 min−1 | [1] |
| k18 | 5min−1 nM−1 | [1] |
| k19 | 1min−1 | [1] |
| k20 | 10 min−1 | [1] |
| k21 | 5min−1 | [1] |
| k22 | 47 min−1 | [1],[4] |
| k23 | 5min−1 | [1],[4] |
| k24 | 345 min−1 | [1],[4] |
| k25 | 5min−1 | [1] |
| k26 | 50 min−1 | [1] |
| k27 | 5min−1 | [1] |
| k28 | 140 min−1 | [1],[5] |
| k29 | 10 min−1 nM−1 | [1] |
| k30 | 1min−1 | [1] |
| k31 | 250 min−1 | [1] |
| k32 | 5min−1 | [1],[5] |
| k33 | 50 min−1 | [1] |
| k34 | 18 min−1 nM−1 | [1,[6]] |
| k35 | 10 min−1 | [1] |