Difference between revisions of "Team:Chalmers-Gothenburg/Theory"
| Line 12: | Line 12: | ||
<p> The receptors share a common structure among all eukaryotic cells, and a major characteristic is, as the name suggests, that the receptor has a transmembrane region which passes through the membrane seven times. The N-terminal of the receptor is located extracellularly and it is responsible for transporting and annealing the protein to the cell membrane. In some cases it also serves as a ligand binding site, although it is more common that the ligands associate with one of the three extracellular loops that reside between the transmembrane sites. The C-terminal of the GPC receptor is a cytosolic region and thus has no ligand associating function whatsoever, instead it has an important role as a mediator between the receptor and the cell or more specifically the G protein that associates with the receptor [1, 2, 3].</p> | <p> The receptors share a common structure among all eukaryotic cells, and a major characteristic is, as the name suggests, that the receptor has a transmembrane region which passes through the membrane seven times. The N-terminal of the receptor is located extracellularly and it is responsible for transporting and annealing the protein to the cell membrane. In some cases it also serves as a ligand binding site, although it is more common that the ligands associate with one of the three extracellular loops that reside between the transmembrane sites. The C-terminal of the GPC receptor is a cytosolic region and thus has no ligand associating function whatsoever, instead it has an important role as a mediator between the receptor and the cell or more specifically the G protein that associates with the receptor [1, 2, 3].</p> | ||
| + | |||
| + | <p> The GPC-receptors as the name indicates are bound to G proteins, as seen in figure 2, within the cell. G proteins are trimeric complexes consisting of three different proteins, called Gα, Gβ, and Gγ. G protein is an abbreviation for guanine nucleotide-binding protein and as the name implies is a protein which binds guanine in the form of GDP or GTP. In a natural state the protein prefers to be bound by GDP, while this is the state the trimeric complex is bound together and has no catalytic activity. During ligand association of the GPC receptor the GDP previously bound to the Gα is replaced with a GTP. While the Gα protein is bound by a GTP the Gβ and Gγ separates as a dimer. In this separate phase the Gβγ or the Gα subunit are free to interact with other molecules, this often leads to the activation of a pathway that ultimately causes a cellular response to an external signal [1, 2, 3].</p> | ||
</html>[[File:ChalmersGothenburgDASTHEORY2.png|700px]]<html> | </html>[[File:ChalmersGothenburgDASTHEORY2.png|700px]]<html> | ||
<p class="figuretext"> Figure 2. A general GPC receptor coupled with a G protein. The figure shows how the activation of the GPC receptor causes the GDP bound to a trimer G protein to be replaced with a GTP, resulting in the splitting of the trimer.</p> | <p class="figuretext"> Figure 2. A general GPC receptor coupled with a G protein. The figure shows how the activation of the GPC receptor causes the GDP bound to a trimer G protein to be replaced with a GTP, resulting in the splitting of the trimer.</p> | ||
| − | |||
| − | |||
| − | |||
<h3>Phereomone response pathway</h3> | <h3>Phereomone response pathway</h3> | ||
Revision as of 01:18, 19 September 2015


Detection and amplification of signal
To detect contaminations in a bioreactor we use a system that utilizes extracellular receptors responding to ligands of the unwanted contaminant. The detection system goes by the name DAS, detection and amplification of signal, and it is based on the pheromone response pathway in Saccharomyces cerevisiae. The pheromone pathway uses a G-protein coupled receptor (GPC-receptor) to detect pheromones of another yeast mating type. When the receptor comes in contact with the correct agonist, which is pheromone α for a-type cells and pheromone a for α-type cells, the receptor then activates a pathway which leads to expression of genes that prepare the yeast for mating. In our system we plan to hijack this system and instead make the yeast signal that a contamination is present.
G protein coupled receptors
GPC receptors, figure 1, also known under the name seven-transmembrane receptors, are among the most common and versatile family of receptors found in eukaryotes. The receptors found in this kind of system are signal mediators through many subjects, such as light, olfactory stimulation, and many other signaling substances commonly found in mammals. They act by activating signal transduction pathways inside the cell, resulting in an intracellular response to an external signal.
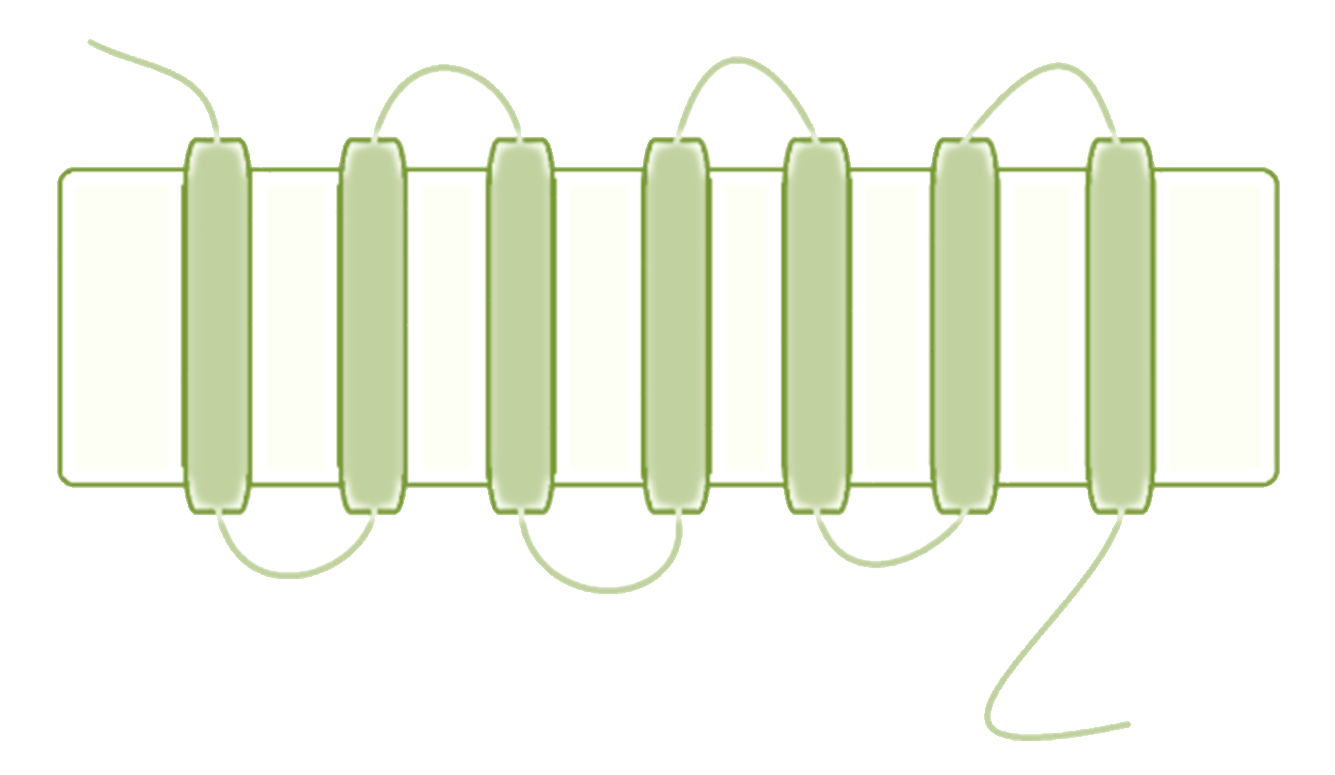
Figure 1. A general GPC receptor. The figure depicts the structure of such a receptor with emphasis on the characteristic transmembrane structure.
The receptors share a common structure among all eukaryotic cells, and a major characteristic is, as the name suggests, that the receptor has a transmembrane region which passes through the membrane seven times. The N-terminal of the receptor is located extracellularly and it is responsible for transporting and annealing the protein to the cell membrane. In some cases it also serves as a ligand binding site, although it is more common that the ligands associate with one of the three extracellular loops that reside between the transmembrane sites. The C-terminal of the GPC receptor is a cytosolic region and thus has no ligand associating function whatsoever, instead it has an important role as a mediator between the receptor and the cell or more specifically the G protein that associates with the receptor [1, 2, 3].
The GPC-receptors as the name indicates are bound to G proteins, as seen in figure 2, within the cell. G proteins are trimeric complexes consisting of three different proteins, called Gα, Gβ, and Gγ. G protein is an abbreviation for guanine nucleotide-binding protein and as the name implies is a protein which binds guanine in the form of GDP or GTP. In a natural state the protein prefers to be bound by GDP, while this is the state the trimeric complex is bound together and has no catalytic activity. During ligand association of the GPC receptor the GDP previously bound to the Gα is replaced with a GTP. While the Gα protein is bound by a GTP the Gβ and Gγ separates as a dimer. In this separate phase the Gβγ or the Gα subunit are free to interact with other molecules, this often leads to the activation of a pathway that ultimately causes a cellular response to an external signal [1, 2, 3].
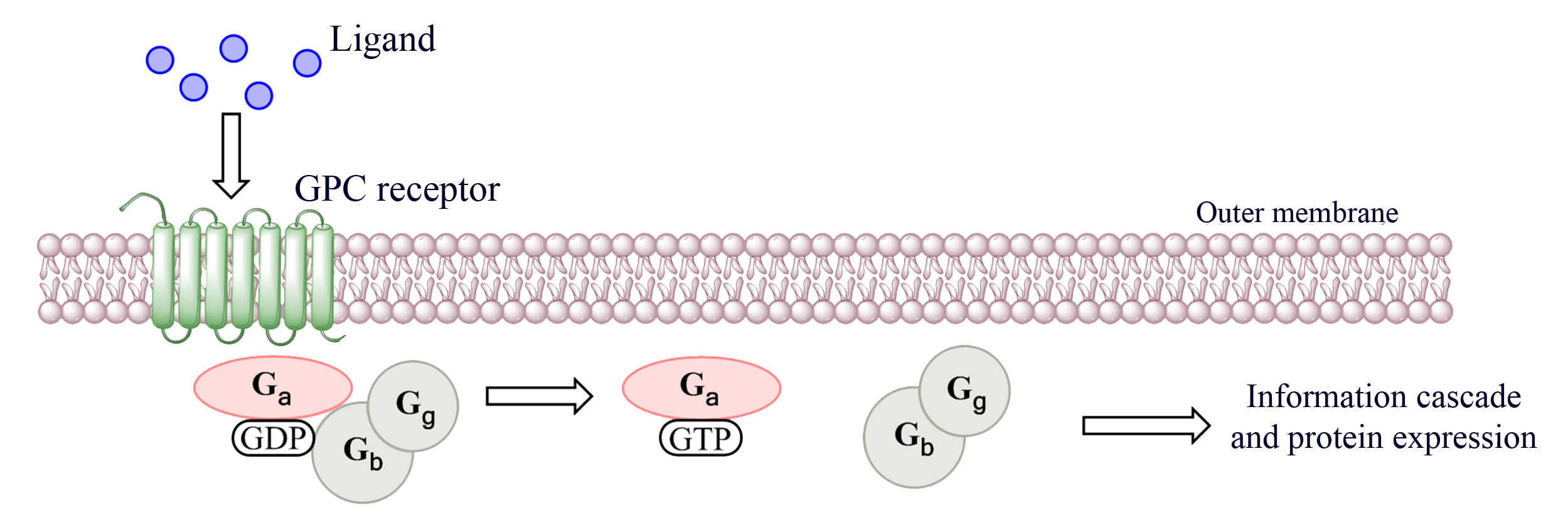
Figure 2. A general GPC receptor coupled with a G protein. The figure shows how the activation of the GPC receptor causes the GDP bound to a trimer G protein to be replaced with a GTP, resulting in the splitting of the trimer.
Phereomone response pathway
The pheromone response pathway in yeast is one of the most studied pathways of eukarya. Due to yeasts important position as a role model organism for eukaryotic cells much attention has been given to the few GPCR pathways that exist in it, the pheromone pathway is one of them. The pathway induces changes such as a cell cycle arrest and conformational changes due to filamentous response genes when a pheromone of a mating partner is located. The following figure depicts a simplified version of the pheromone response pathway.
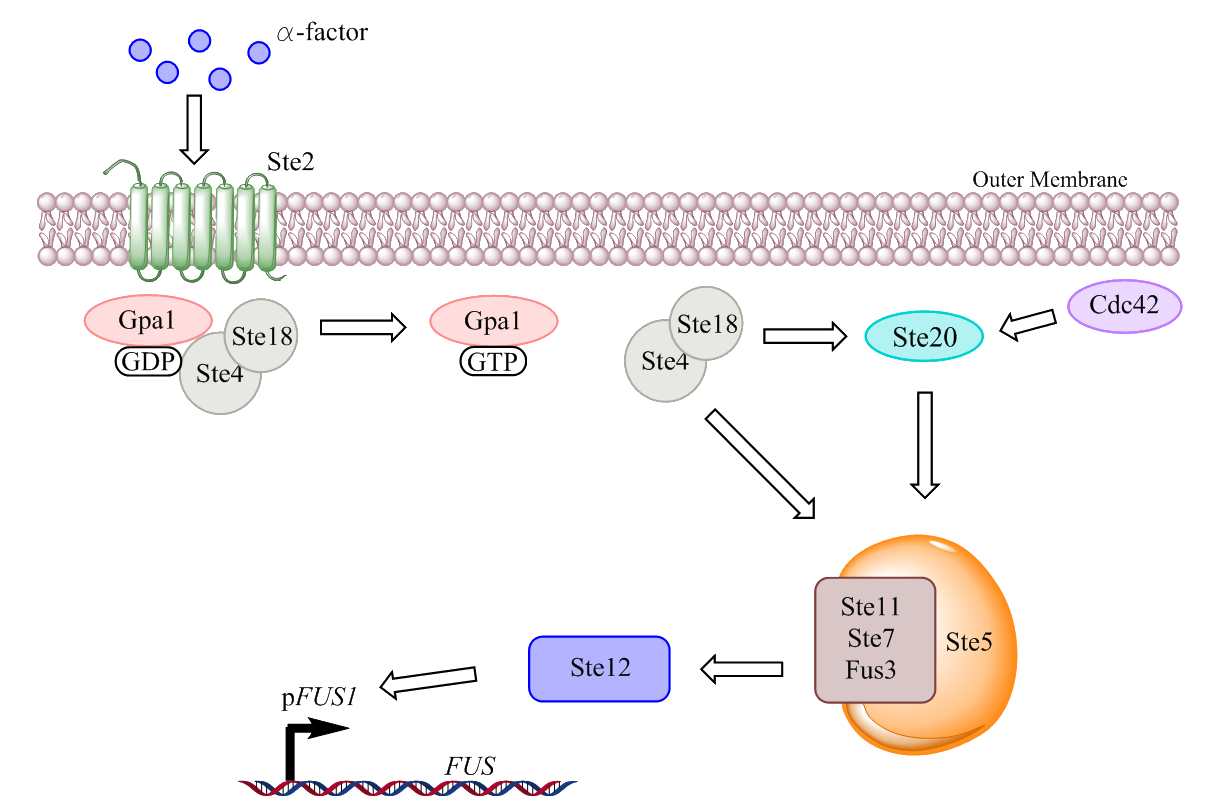
Figure 3. A simplified visualization of the pheromone response pathway in S. cerevisiae. The figure shows how the signaling cascade is activated by an agonist ligand, resulting in the induced expression of certain genes.
The figure depicts an a-type yeast cell containing the receptor Ste2, this receptor interacts with the pheromone from the α-type yeast cell called α-factor. The receptor Ste2 is in turn intracellularly bound to the three G-protein units α, γ, and β named Gpa1, Ste18, and Ste4 respectively. During ligand association the agonist pheromone produces a conformational change in the receptor structure, exposing parts of the receptor which catalyzes the exchange of the GDP bound to the intracellular α-subunit to a GTP. When the diphosphate is replaced with a triphosphate the conformation of the trimer changes, and the β- and γ-subunits separate as a dimer from the α-subunit.
This Gβγ dimer interacts with two effectors called Ste20 and Ste5. Ste20 is a PAK (p21-activated protein kinase) and its kinase activity is enabled when it is bound to the G protein Cdc42. Ste5 is a scaffold protein which attracts and binds the three proteins Ste11, Ste7, and Fus3 which are MEKK/MAP3K (Mitogen activated protein kinase kinase kinase), MEK/MAP2K (Mitogen activated protein kinase kinase), and MAPK (Mitogen activated protein kinase) respectively. When both Ste5 and Ste20 bind to the Gβγ dimer it allows for the activated Ste20 kinase to phosphorylate the Ste11 MEKK thus triggering a MAPK cascade. This phosphorylation cascade ends in the release of activated Fus3, which in turn is transported to the nucleus where it activates the transcription factor Ste12 [3].
Detection system
In our system, the natural receptor (Ste2 from a-cells) has been replaced with a fusion receptor consisting partially of the natural Ste2 receptor but mostly a pheromone receptor called Mam2 originating from the yeast Schizosaccharomyces pombe. Due to both of those receptors being GPC-receptors with a similar Gα subunit the fusion receptor is capable of interacting with the G-protein of the pheromone response pathway in Saccharomyces cerevisiae. Depicted below is the modified system we use for detection:
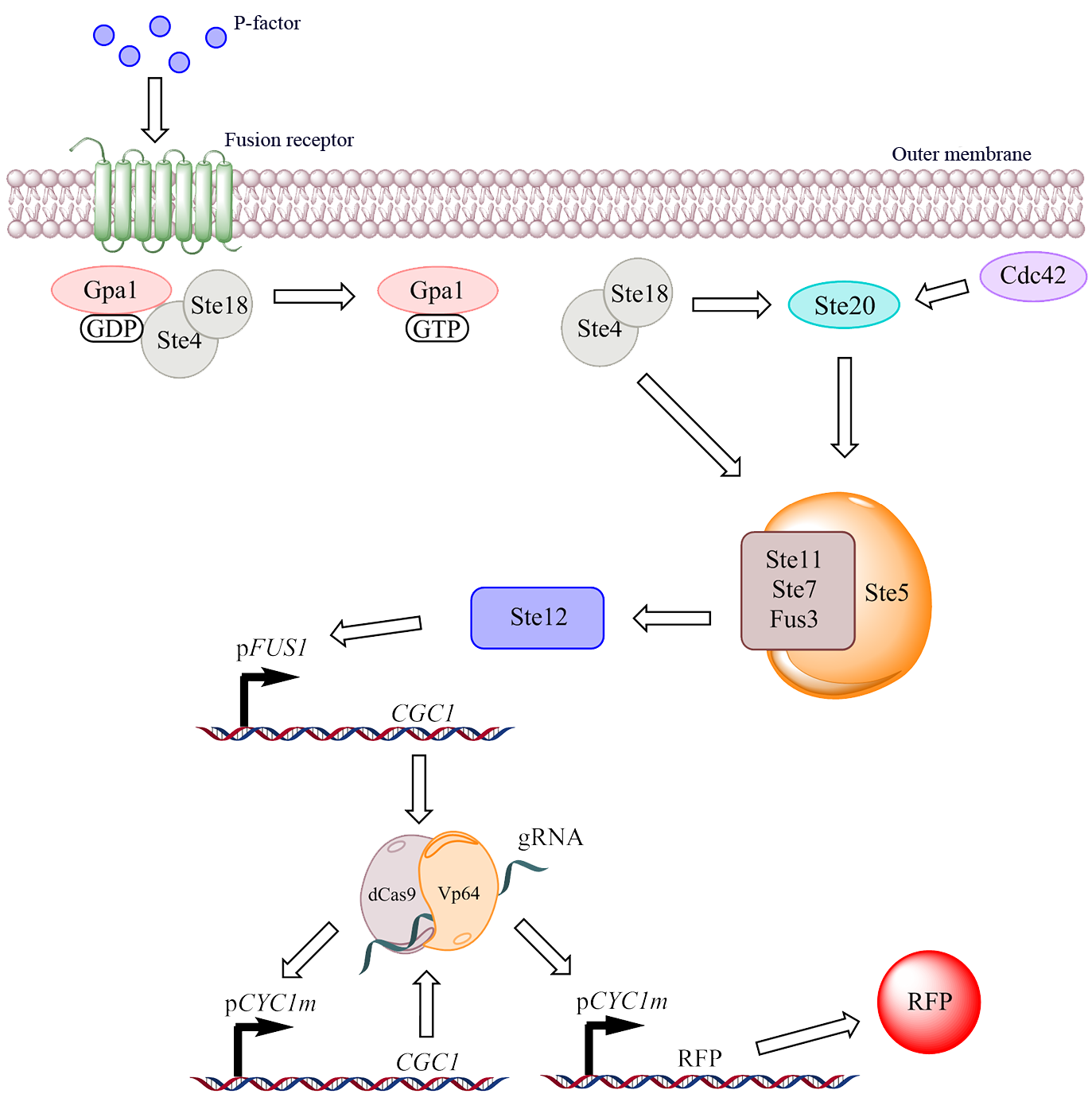
Figure 4. The DAS system as a whole. The figure depicts how the natural GPC receptor of the pheromone pathway has been replaced with a fusion receptor that is activated by an alternative agonist ligand, the resulting signaling cascade results in a signal amplification system that gives an output signal in the form of fluorescent proteins.
Fusion receptor
The fusion receptor contains the N-terminus signal peptide of the Ste2 receptor so that the receptor is correctly transported and integrated into the cell membrane of the S. cerevisiae cell wall. The intermediate region and the C-terminus of the receptor is replaced with the equivalent parts of the Mam2 receptor, which makes the receptor capable of interacting with the correct ligand. Since the GPC-receptor is mostly dependent on the extracellular loops in regard of what ligand it associates with it is possible to create a wide array of receptors simply by replacing this region in the receptor [4]. There are many receptors to be found in the eukaryotes, each with its own specificity [5].
Amplification
In order to produce a strong output signal when a contaminant is detected we have decided to use intracellular amplification to strengthen the signal. To achieve this in an efficient manner, while holding on to the work previously done in the iGEM competition we decided to use the dCas9-Vp64 complex that was used the previous year by Team Gothenburg. By linking an activation domain to a deactivated version of the CRISPR associated protein, dCas9, one obtains a transcription factor guided by the same means as the popular CRISPR system. By creating a promoter with a binding site for a specific RNA sequence and expressing this RNA to act as a guide RNA (gRNA for short) for the dCas9 protein it is possible to make an incredibly precise tool for gene expression.
Vp64 is a viral protein complex consisting of four Vp16 activation domains, originating from the herpes virus, which when combined form the strong activation domain called Vp64. By coupling this activation domain to a DNA binding protein such as dCas9 a strong, activating transcription factor is created [6].
However, the expression of a new transcription factor in itself, even though it induces a strong expression, is not guaranteed to produce a strong final signal, therefore we decided to amplify the amount of transcription factor through a feed-back activation loop, where we coupled the expression of the transcription factor (dCas9-Vp64) to the promoter that it specifically binds to, pCYC1m. The results of this coupling should be a heightened amount of transcription factor in the cell, allowing for close to the maximum expression of the signal substance, in our case RFP. The dCas9-Vp64 gene is transcribed together with the gRNA as a single mRNA molecule (see figure 5), but thanks to the incorporation of ribozymes these two RNA parts are separated [7].
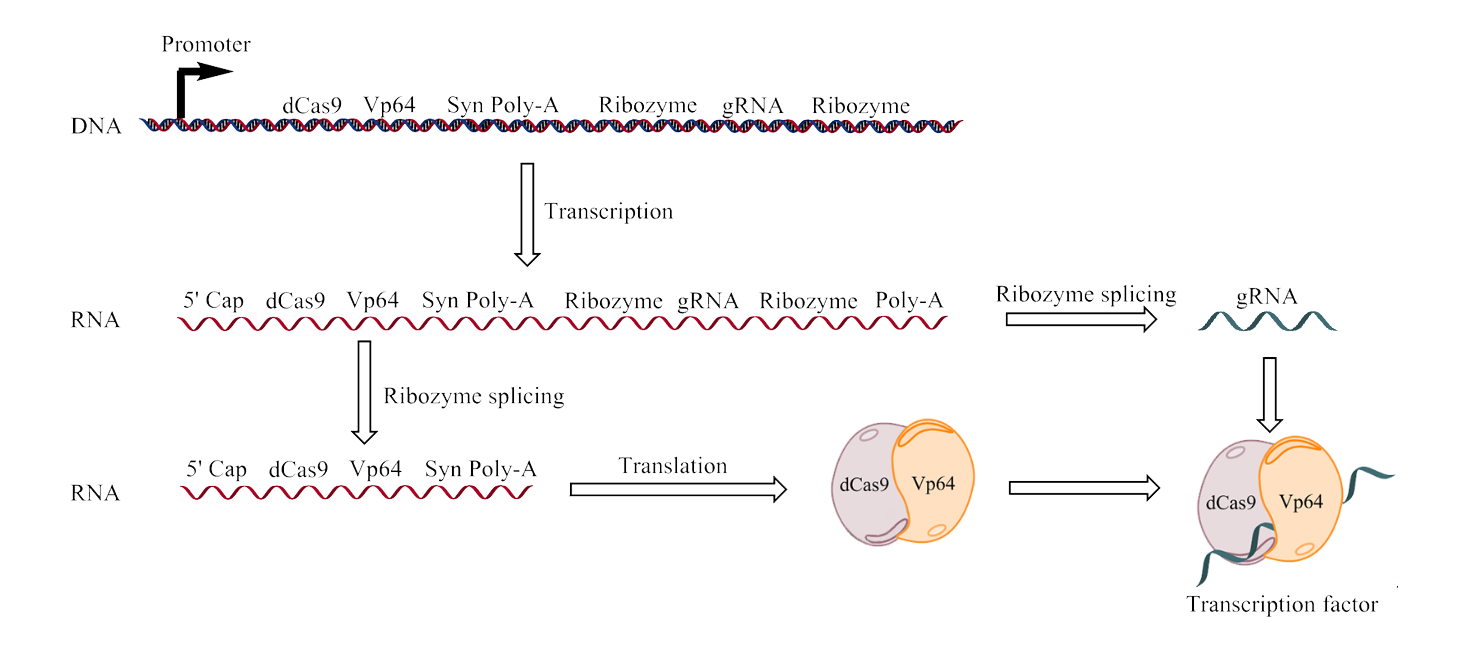
Figure 5. A close up of the CGC1 gene, the figure depicts the transcription of the CGC1 gene and the following ribozyme splicing leading up to the creation of a gRNA and the transcription of the dCas9-Vp64 complex. When these two components are combined they create a transcription factor.
References:
[1] Trzaskowski, B., Latek, D., Yuan, S., Ghoshdastider, U., Debinski, A., & Filipek, S. (2012). Action of molecular switches in GPCRs - theoretical and experimental studies. Current Medicinal Chemistry, 19(8), 1090-1109. doi:10.2174/092986712799320556
[2] Bockaert, J., & Philippe Pin, J. (1999). Molecular tinkering of G protein-coupled receptors: An evolutionary success. The EMBO Journal, 18(7), 1723-1729. doi:10.1093/emboj/18.7.1723
[3] Bardwell, L. (2004). A walk-through of the yeast mating pheromone response pathway. Peptides, 25(9), 1465-1476. doi:10.1016/j.peptides.2003.10.022
[4] Fukutani, Y., Nakamura, T., Yorozu, M., Ishii, J., Kondo, A., & Yohda, M. (2012). The N‐terminal replacement of an olfactory receptor for the development of a Yeast‐based biomimetic odor sensor. Biotechnology and Bioengineering, 109(1), 205-212. doi:10.1002/bit.23327
[5] Hopkins, A. L., Overington, J. P., & Al-Lazikani, B. (2006). How many drug targets are there? Nature Reviews Drug Discovery, 5(12), 993-996. doi:10.1038/nrd2199
[6] Maeder, M. L., Linder, S. J., Cascio, V. M., Fu, Y., Ho, Q. H., & Joung, J. K. (2013). CRISPR RNA-guided activation of endogenous human genes. Nature Methods, 10(10), 977-979. doi:10.1038/nmeth.2598
[7] Dower, K., Kuperwasser, N., Merrikh, H., & Rosbash, M. (2004). A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA, 10(12), 1888-1899. doi:10.1261/rna.7166704
DNA-extension dependent DNA-repair
Deinococcus radiodurans is an extremophilic bacterium with an extraordinary ability to survive vast amounts of DNA-damaging radiation [1, 3]. It is one of the most radiation-resistant organisms known and has even been listed in the Guinness Book of World Records as “the world’s toughest bacterium” [6]. Many DNA damaging agents such as γ-radiation, which is a particularly lethal form of ionizing radiation, induce double-stranded breaks (DSBs) in the genome during exposure. These DSBs are very difficult to repair since there is no template to direct DNA repair as with single-stranded breaks. The amount of ionizing radiation needed to kill 63% of the cells in a population is called the D37 dose. For D. radiodurans, the D37 dose is 6000 Gy. This amount of radiation is estimated to induce 275 DSBs and 3000 single-stranded breaks per genome. For comparison, the introduction of less than 10 DSBs per genome is lethal to Escherichia coli as well as to most other species. [2, 5]
How can D. radiodurans be so resistant?
This remarkable resistance is due to the extremely proficient DNA repair processes of D. radiodurans. Yet little is known about the precise mechanisms behind this efficient repair. Most, if not all, of the typical bacterial DNA-repair proteins are found in D. radiodurans. So how come that Deinococcus radiodurans is so resistant to ionizing radiation? [4] One possibility is that it utilizes repair mechanisms that are fundamentally different from all other bacteria. Another possibility is that it uses the same repair systems as all the others, but in a more effective way. [3, 4] It appears as if D. radiodurans is able to limit DNA degradation and restrict diffusion of DNA fragments that are produced when irradiated. But what mechanisms underlie these features is still being debated. Both non-homologous end-joining (NHEJ) and extended synthesis-dependent strand annealing (ESDSA) have been suggested. [5]
NHEJ is claimed to be a useful process for repairing double-stranded breaks in situations where strand ends might not be able to freely diffuse away from each other, such as in condensed chromosomes. NHEJ is present in other bacteria and certain enzymes known to be involved in NHEJ are present in D. radiodurans. [5, 9] However, this mechanism is error-prone which doesn’t consort with the accurate genome repair seen in D. radiodurans. [5, 10]. In ESDSA, single-stranded overhangs with overlapping homologies on shattered chromosomes are used both as primers and templates for synthesis of complementary strands. The elongated complementary strands can then reestablish chromosomal integrity with high precision. This rather extensive DNA repair synthesis is dependent on the activity of the RecA protein. However, it doesn’t seem to be able to explain all the means behind D. radiodurans’s incredible ability to withstand vast amounts of radiation and oxidative stress. [7, 8] The most likely scenario is that several different pathways act alongside each other in D. radiodurans where some are more similar to the pathways used in other organisms, and some are more specific for D. radiodurans.
Repair in Our project
Our plan is therefore to identify the pathway that is the most specific for D. radiodurans and the specific proteins participating in that pathway. We will then insert these into S. cerevisiae to implement a unique system for post-UV radiation resistance (PUR) in the yeast to increase its resistance to UV irradiation, which will be used in the bioreactor to eliminate contaminants. The RecA system found in D. radiodurans is unique when compared to other organisms, since it can repair DNA breaks with both sticky and blunt ends. [11] This process involves the proteins RecA, RecQ, RecD2, PprA and DrSSB, and also RecJ, which is often found in the RecF pathway in other organisms. This exonuclease is used both in the RecFOR pathway in D. radiodurans as well as in the RecA pathway to create single-stranded DNA ends to which the other enzymes can associate. Since D. radiodurans is polyploid, the repair system is based on an organism with multiple genome copies, which is why a diploid yeast is going to be used. Also, since the proteins need to be transported into the nucleus to be able to associate with the DNA, an NLS sequence will be added to each protein.
The repair mechanism
At the presence of DNA breaks, the exonuclease RecJ in our system will start the repair process by hydrolyzing the 5’-terminal strand of both fragments, generating two single-stranded fragments. To avoid degradation and formation of hairpin structures in the exposed single DNA strands, DrSSB will attach to the DNA strands. Apart from stabilizing the DNA, SSB can then interact with the RecA enzyme to initiate DNA strand exchange. The RecA enzyme will introduce complementary sequences to the ssDNA, which can be found among the additional genomes in the polyploid organism. RecQ also accompanies RecA, this 3’-5’ helicase will unwind the two strands in the template brought by RecA to allow for a DNA polymerase to associate with the DNA strands. DrSSB will bind to these single DNA strands as well, to avoid self-annealing and protect the strands from degradation. The DNA polymerase already present in yeast will attach to the template and start elongating the sequence of the damaged gene. Following the elongation, the 5’-3’ helicase RecD2 will unwind and release the template from the gene fragment, which hybridizes to the complementary sequence created by the DNA polymerase. After the helicase has released the template, the DNA polymerase can associate once more to the gene fragment, to fully elongate the remaining sequence. The polymerase is accompanied by PprA, a ligase-inducing enzyme, to fully repair the gaps in the sequence.
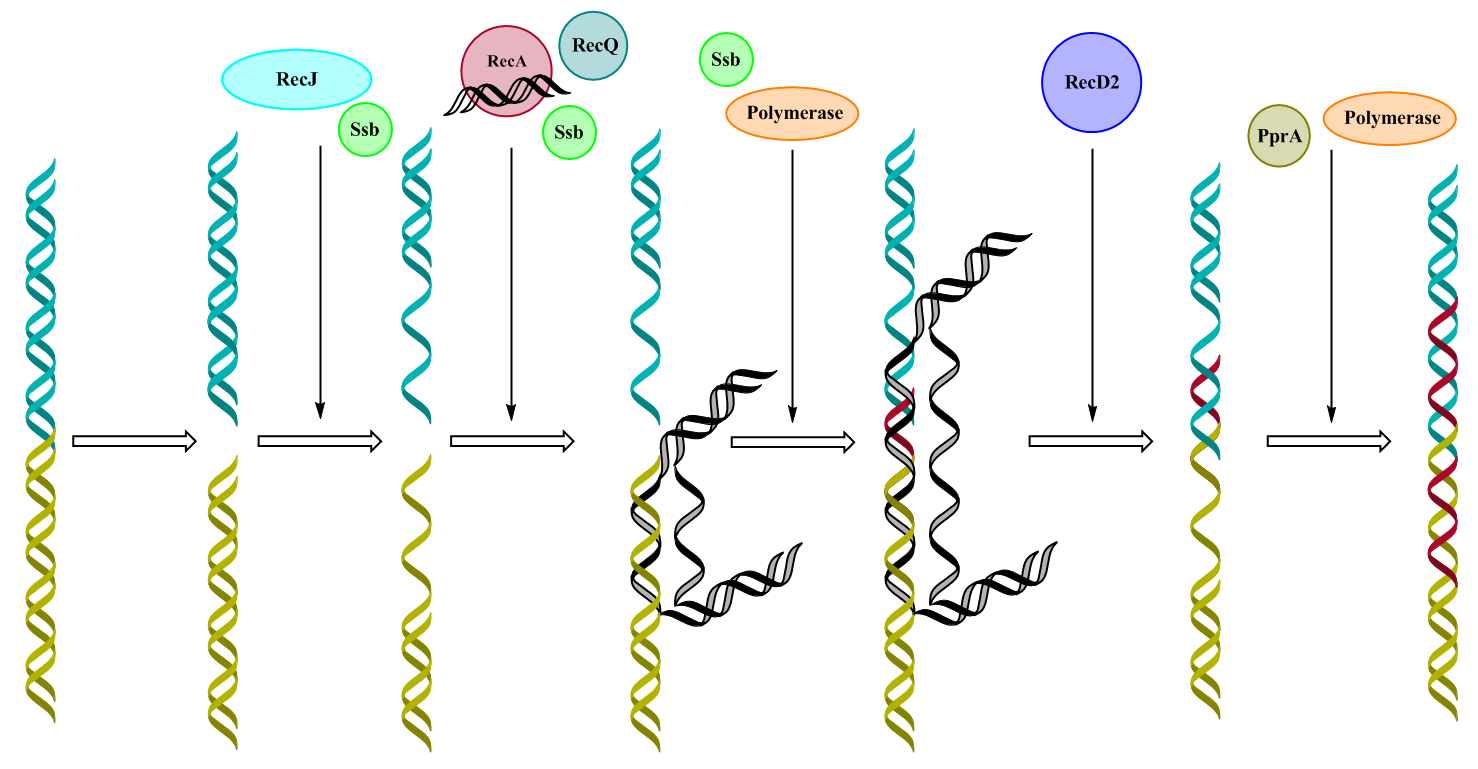
Figure 6. The PUR system as a whole. The figure depicts how the incorporated enzymes from the RecA system in D. radiodurans collaborate with DNA repair enzymes naturally present in yeast, such as the DNA polymerase, to repair a double-stranded break.
References:
[1] Minton, K. W. (1996). Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutation Research/DNA Repair, 363(1), 1-7.
[2] Killoran, M. P., & Keck, J. L. (2006). Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. Journal of Biological Chemistry, 281(18), 12849-12857.
[3] Battista, J. R. (1997). Against all odds: the survival strategies of Deinococcus radiodurans. Annual Reviews in Microbiology, 51(1), 203-224.
[4] Battista, J. R., Earl, A. M., & Park, M. J. (1999). Why is Deinococcus radiodurans so resistant to ionizing radiation?. Trends in microbiology, 7(9), 362-365.
[5] Cox, M. M., & Battista, J. R. (2005). Deinococcus radiodurans—the consummate survivor. Nature Reviews Microbiology, 3(11), 882-892.
[6] Sarah E. DeWeerdt, SDW. (2002) The World’s Toughest Bacterium – Deinococcus radiodurans may be a tool for cleaning up toxic waste and more. Genome News Network. July 5th 2002. http://www.genomenewsnetwork.org/articles/07_02/deinococcus.shtml (September 12th 2015).
[7] Zahradka, K., Slade, D., Bailone, A., Sommer, S., Averbeck, D., Petranovic, M., ... & Radman, M. (2006). Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature, 443(7111), 569-573.
[8] Slade, D., Lindner, A. B., Paul, G., & Radman, M. (2009). Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell, 136(6), 1044-1055.
[9] Englander, J., Klein, E., Brumfeld, V., Sharma, A. K., Doherty, A. J., & Minsky, A. (2004). DNA toroids: framework for DNA repair in Deinococcus radiodurans and in germinating bacterial spores. Journal of bacteriology, 186(18), 5973-5977.
[10] Daly, M. J., & Minton, K. W. (1996). An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. Journal of bacteriology, 178(15), 4461-4471.
[11] Cox, M. M., Keck, J. L., & Battista, J. R. (2010). Rising from the Ashes: DNA Repair in Deinococcus radiodurans. PLoS Genet, 6(1), e1000815.
The repair proteins
RecA
RecA and RecA-like proteins have been found throughout the phylogenetic tree and their structures are often conserved between species. They play a critical role in the repair processes that require homologous DNA pairing and recombination. The RecA protein from D. radiodurans is essential for the remarkable radiation resistance of the organism and is in many aspects a functional homolog to the protein found in E. coli. [6] The protein from D. radiodurans has been shown to be able to, at least partially, complement the mutated or deleted homolog in E. coli and vice versa [4, 7]. RecA is the protein responsible for promoting DNA strand exchange upon DNA damage in D. radiodurans.
RecA was first discovered in a DNA damage-sensitive strain of D. radiodurans which was found to be defective in a gene homologous to the RecA gene in E. coli. When transformed with a chromosomal DNA fragment that contained the deinococcal RecA gene from wild-type, the DNA damage resistance in the mutated strain was fully restored. [1] Even though the complete DSB rejoining in D. radiodurans is RecA-dependent, the initial repair is not. D. radiodurans is able to rejoin many DSBs in a sequence-specific manner for the first 1.5 hours, as seen in RecA mutants, before the RecA-dependent homologous recombination is initiated. [3] RecA is thus not detectable in the organism until DNA damage occurs and DNA damage repair has begun [2].
DNA strand exchange
The common pathway for DNA strand exchange, which is promoted by EcRecA and many of its homologs, is initiated by the formation of a filament on ssDNA, followed by uptake of the duplex substrate. These proteins thus target single-stranded gaps and tails. The RecA protein from D. radiodurans performs the exact inverse of this pathway. [5] There are three reasons for this. (i) DrRecA binds preferentially to dsDNA rather than ssDNA and rather initiates DNA strand exchange with a filament bound to dsDNA. [5, 6] (ii) Heterologous DNA strongly inhibits the DNA strand exchange by DrRecA, unlike EcRecA. (iii) In a DNA strand exchange reaction between a short linear dsDNA and a long circular ssDNA, the amount of DrRecA protein required for optimal reaction is stoichiometric with the duplex DNA and is unaffected by changes in ssDNA concentration, where the opposite is true for EcRecA. [5] One common aspect though is that DNA strand exchange is greatly facilitated by SSB proteins for both DrRecA and EcRecA [6].
The significance of using the inverse pathway may be related to the absence of the recB and recC genes in the deinococcal genome. By operating the inverse pathway, RecBC is not needed to prepare single-stranded tails since DrRecA binds directly to dsDNA to promote repair of DSBs. [5]
References:
[1] Gutman, P. D., Carroll, J. D., lan Masters, C., & Minton, K. W. (1994). Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene, 141(1), 31-37.
[2] Carroll, J. D., Daly, M. J., & Minton, K. W. (1996). Expression of recA in Deinococcus radiodurans. Journal of bacteriology, 178(1), 130-135.
[3] Daly, M. J., & Minton, K. W. (1996). An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. Journal of bacteriology, 178(15), 4461-4471.
[4] Narumi, I., Satoh, K., Kikuchi, M., Funayama, T., Kitayama, S., Yanagisawa, T., ... & Yamamoto, K. (1999). Molecular analysis of the Deinococcus radiodurans recA locus and identification of a mutation site in a DNA repair-deficient mutant, rec30. Mutation Research/DNA Repair, 435(3), 233-243.
[5] Kim, J. I., & Cox, M. M. (2002). The RecA proteins of Deinococcus radiodurans and Escherichia coli promote DNA strand exchange via inverse pathways. Proceedings of the National Academy of Sciences, 99(12), 7917-7921.
[6] Kim, J. I., Sharma, A. K., Abbott, S. N., Wood, E. A., Dwyer, D. W., Jambura, A., ... & Cox, M. M. (2002). RecA Protein from the extremely radioresistant bacterium Deinococcus radiodurans: expression, purification, and characterization. Journal of bacteriology, 184(6), 1649-1660.
[7] Schlesinger, D. J. (2007). Role of RecA in DNA damage repair in Deinococcus radiodurans. FEMS microbiology letters, 274(2), 342-347.
RecQ
RecQ helicases are key enzymes in genome maintenance that participate in DNA replication, recombination, and repair. This multifunctional enzyme initiates homologous recombination and suppresses illegitimate recombination. RecQ is a member of the SF1 family of DNA helicases, which are highly conserved from bacteria to eukaryotes.[1,2] In vitro, RecQ helicases are able to exchange and unwind a wide variety of DNA substrates, including G4 DNA and three- or four-stranded DNA structures such as D-loops and Holliday junctions. It has also been suggested that RecQ generates an initiating signal that can recruit RecA for SOS induction and recombination at stalled replication forks.[3]
In E.coli, null mutations at the recQ locus result in a significant reduction of the efficiency of homologous recombination as well as an increase in sensitivity to UV radiation [6]. Similar deletions of RecQ in D. radiodurans resulted in a low doubling rate and sensitivity to hydrogen peroxide, γ-radiation, UV-light and mitomycin [1,5]. In addition to this, the expression of certain genes involved in iron homeostasis, antioxidant system, electron transport, and energy metabolism were significantly altered, indicating that RecQ has pleiotropic functions in the organism and thereby contributes to D. radiodurans’s astonishing ability to withstand stresses. A study conducted by Chen et al. discovered that upon deletion of DrRecQ, ROS accumulation increased and Fe and Mn metabolisms were altered.[5] Since RecQ helicases have such diverse cellular functions, they require specialization and coordination of multiple protein domains that integrate catalytic functions between both DNA-protein and protein-protein interactions [4].
Domains
Most RecQ helicases have three conserved domains. These are designated as the helicase, RecQ-conserved (RQC), and Helicase and RNase D C-terminal (HRDC) domains in order from N- to C-terminus. The helicase region in the RecQ enzyme unwinds DNA with 3’→5’ polarity with respect to the DNA strand to which the enzyme is bound by using the energy from ATP hydrolysis. The RQC domain, which is unique to RecQ helicases, might have a role in regulating specific protein–protein interactions and adjusting the enzyme’s binding affinity to DNA. [1] HRDC domains are involved in structure-specific nucleic acid binding and assist in targeting RecQ proteins to particular DNA structures.[1,2] In Deinococcus radiodurans, DR1289 is a special member of RecQ family [1]. The structure of the domains in this enzyme is similar to RecQ helicases from other organisms, except that it encodes three HRDC domains whereas most only encode one.[1,2,5]
If the HRDC domains have been inactivated, the RecQ enzyme can usually function as a DNA-dependent ATPase and helicase, but it often has altered DNA binding activities. Neither the helicase domain nor the three HRDC domains alone are capable of fully restoring the DNA damage resistance compared to a wild type strain. Nevertheless, the helicase and all three HRDC domains are crucial for complete DNA damage resistance in D. radiodurans. As each HRDC domain is sequentially removed from the C-terminus, D. radiodurans displays a gradual decrease in DNA damage resistance. The unique tandem C-terminal arrangement of the HRDC domains is important for DNA damage resistance.[1,2]
The three HRDC domains in D. radiodurans have evolved to function in DNA binding and regulation. Together they increase the efficiency of the unwinding and ATPase activities of the helicase domain and are important for optimal helicase activity in vitro – removal of all three domains produces and enzyme with a considerably weakened ability to bind DNA.[1,2] However, each domain has a distinct effect on these activities as well as the structure-specific DNA binding of the enzyme. The N-terminal-most HRDC domain is critical for high affinity DNA binding and for efficient unwinding of DNA, whereas the more C-terminal HRDC domains lessen the DNA binding affinity and the ATPase rate. These two domains also possess more complex roles in structure-specific DNA unwinding. [2] The C-terminal-most HRDC domain has an unusual electrostatic surface that regulate structure-specific DNA binding and help direct DrRecQ to specific recombination/repair sites. This domain features a concentrated patch of acidic residues that provides a surface for inter-domain interactions which direct the enzyme to bind to certain DNA structures.[4]
Although the HRDC domains do not possess any helicase activity, expression of only the C-terminal portion of Dr1289 in a deletion mutant has been shown to be able to partially restore resistance to DNA damaging agents [1]. Another study showed that, unlike single HRDC RecQ helicases, DrRecQ does not simply use its HRDC domains as supplementary DNA binding elements [2]. These activities suggest that the three tandem HRDC domains have evolved to perform an essential function in DNA repair with more complex regulatory roles. The HRDC domains may increase the number of substrates to the enzyme or aid in substrate selection, aside from regulating the level of activity of the helicase.[1,2] Furthermore, because the C-terminal-most HRDC domain alone can partially restore resistance, the recognition of substrate by the tandem HRDCs may recruit other repair factors. For instance, RecQ and RecA interact in E. coli recombination, and DrRecQ may function analogously in DNA repair.[1] Additional potential roles of DrRecQ HRDC domains include acting as sites for protein-protein interactions that could target the enzyme to damaged DNA structures and/or provide a cellular mechanism for regulating the actions of the HRDC domains on DrRecQ in different contexts.[2]
Specific interactions
SSB proteins prevent the formation of hairpins in single-stranded DNA and thereby stimulate RecQ helicase-promoted unwinding of single-stranded DNA substrates. The unwinding rate of Dr1289 has been shown to increase in the presence of DrSSB, which has a species-specific effect on the enzyme.[1]
References:
[1] Huang, L., Hua, X., Lu, H., Gao, G., Tian, B., Shen, B., & Hua, Y. (2007). Three tandem HRDC domains have synergistic effect on the RecQ functions in Deinococcus radiodurans. DNA repair, 6(2), 167-176.
[2] Killoran, M. P., & Keck, J. L. (2006). Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. Journal of Biological Chemistry, 281(18), 12849-12857.
[3] Huang, L. F., Hua, X. T., Lu, H. M., Gao, G. J., Tian, B., Shen, B. H., & Hua, Y. J. (2006). Functional analysis of helicase and three tandem HRDC domains of RecQ in Deinococcus radiodurans. Journal of Zhejiang University Science B, 7(5), 373-376.
[4] Killoran, M. P., & Keck, J. L. (2008). Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ. Nucleic acids research, 36(9), 3139-3149.
[5] Chen, H., Huang, L., Hua, X., Yin, L., Hu, Y., Wang, C., ... & Hua, Y. (2009). Pleiotropic effects of RecQ in Deinococcus radiodurans. Genomics, 94(5), 333-340.
[6] Nakayama, K., Irino, N., & Nakayama, H. (1985). The recQ gene of Escherichia coli K12: molecular cloning and isolation of insertion mutants. Molecular and General Genetics MGG, 200(2), 266-271.
SSB
For DNA replication, recombination and repair to function, e.g. when DNA damage occurs, the dsDNA must be unwound for the necessary enzymes to be able to access the DNA strands. This process is not without risk, ssDNA is prone to chemical and nucelolytic attacks that can produce breaks that are difficult to repair. ssDNA can also self-associate, which inhibits many genome maintenance enzymes. Cells have therefore evolved a specialized class of proteins (ssDNA-binding proteins, or SSBs) that associate with ssDNA, independently of their sequences, to help preserve the ssDNA intermediates and protect them from degradation. These proteins are also known for associating with many different types of genome maintenance enzymes. [1, 4, 6]
Structure
One thing that all SSBs have in common is that they use a conserved domain called an oligonucleotide/oligosaccharide-binding (OB) fold to bind ssDNA. The majority of all bacterial SSBs function as homotetramers, where each monomer contributes a single OB fold. [3] In contrast, SSB from D. radiodurans contains two such folds per monomer. It also functions as a homodimer rather than as a tetramer. [2, 4, 5] The major advantage of linking two OB folds together is that the two OB domains can evolve separately. The asymmetry seen in DrSSB indicates that each domain has developed into possessing specialized roles. [3]
Another common feature is that the last 10 amino acids at the C-terminus are all highly acidic and very well conserved. These C-terminal tails are suspected to act as binding sites for other proteins involved in genome maintenance. An acidic C-terminus is found in the C-terminal tails in DrSSB as well, but unlike the tetrameric SSBs DrSSB only contains two such tails. [2, 3]
Binding
The OB domains of DrSSB bind ssDNA by using a combination of aromatic residues that stack against nucleotide bases, electrostatic, and polar interactions, such as positively charged residues that form ionic interactions with the DNA backbone [3, 5]. The extended surface to which ssDNA binds on the protein is similar to that used by tetrameric enzymes. Although the general binding path is conserved, there are distinct interaction surfaces present in DrSSB that indicate that species-specific binding to ssDNA occurs. [5]
Radiation tolerance
In a study performed by George et al., upon treatment with heavy ionizing radiation, DrSSB protein levels rose significantly in vivo as early as an hour after the treatment. The accumulation of the protein had increased to maximal levels after 3 hours, and the elevated levels persisted for several hours afterwards. The localization of SSB also changed dramatically during the experiment. Immediately after radiation, most SSBs were found in the nucleoid, but as time passed the proteins started to dissipate through the cytosol. This radiation-dependent accumulation suggests that SSB might be important for the robust repair mechanisms in D. radiodurans. [5]
Lockheart et al. recently reported that SSB is essential for cell survival. A deletion of the drssb gene is lethal, and a lowered expression of DrSSB affects ionizing radiation tolerance, ultraviolet radiation tolerance, and general growth capabilities. The fact that ultraviolet and ionizing radiation produce different types of DNA damage highlights that DrSSB appears to be central to all DNA repair pathways, regardless of the type of damage involved. [6]
References:
[1] Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M., & Keck, J. L. (2008). SSB as an organizer/mobilizer of genome maintenance complexes. Critical Reviews in Biochemistry and Molecular Biology, 43(5), 289-318.
[2] Eggington, J. M., Haruta, N., Wood, E. A., & Cox, M. M. (2004). The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC microbiology, 4(1), 2.
[3] Bernstein, D. A., Eggington, J. M., Killoran, M. P., Misic, A. M., Cox, M. M., & Keck, J. L. (2004). Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proceedings of the National Academy of Sciences of the United States of America, 101(23), 8575-8580.
[4] Witte, G., Urbanke, C., & Curth, U. (2005). Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic acids research, 33(5), 1662-1670.
[5] George, N. P., Ngo, K. V., Chitteni-Pattu, S., Norais, C. A., Battista, J. R., Cox, M. M., & Keck, J. L. (2012). Structure and cellular dynamics of Deinococcus radiodurans single-stranded DNA (ssDNA)-binding protein (SSB)-DNA complexes. Journal of Biological Chemistry, 287(26), 22123-22132.
[6] Lockhart, J. S., & DeVeaux, L. C. (2013). The essential role of the Deinococcus radiodurans ssb gene in cell survival and radiation tolerance.
RecJ
DSB repair is initiated by degradation of the 5’-terminal strand by an exonuclease in coordination with a helicase. The recombinase enzyme RecA is then able to carry out a strand exchange with a homologous DNA duplex. This process is called extended synthesis dependent strand annealing (ESDSA) in Deinococcus radiodurans. The degradation of the 5’-terminal strand is most often dependent on the RecBCD helicase or on a related enzyme with similar activity. The D. radiodurans genome lacks a homologue for this protein, so an alternative pathway has been suggested. When RecBCD is absent in E. coli, the RecF pathway is an alternative pathway for recombination and DNA repair. D. radiodurans may also rely on enzymes of this pathway for DSB repair. [1, 2]
The ssDNA-specific nuclease RecJ is found in most bacteria and is thought to process damaged DNA ends in the RecF (or RecFOR) pathway. It is a 5’→ 3’ ssDNA exonuclease which is unable to process dsDNA. [2] The protein requires a metal ion for activity, the most common regulator of its activity is Mn2+ followed by Mg2+, both of which are present in D. radiodurans. Furthermore, DrSSB has been shown to stimulate DrRecJ exonuclease activity as well. [1, 2]
Mutations
Mutants with reduced or deleted RecJ gene copy grow more slowly and are more sensitive to UV and γ-radiation and hydrogen peroxide, suggesting an important role for RecJ in DNA repair in D. radiodurans. [1, 2] One possibility as to why these mutations affect D. radiodurans in such a negative way could be because of the lack of the RecBC genes in the deinococcal genome. Only when the RecBCD gene has been inactivated in E. coli is the RecF pathway active, and studies have shown that RecBC-RecJ double mutants are lethal in certain organisms. [1]
References:
[1] Cao, Z., Mueller, C. W., & Julin, D. A. (2010). Analysis of the recJ gene and protein from Deinococcus radiodurans. DNA repair, 9(1), 66-75.
[2] Jiao, J., Wang, L., Xia, W., Li, M., Sun, H., Xu, G., ... & Hua, Y. (2012). Function and biochemical characterization of RecJ in Deinococcus radiodurans. DNA repair, 11(4), 349-356.
RecD2
DSBs are thought to be repaired through homologous recombination, a process that depends on the RecA and RecBCD enzymes in most organisms [5]. In general, the RecBCD enzyme processes the broken DNA ends through combined nuclease and helicase activities and loads the RecA protein onto the resulting single DNA strand. The single-stranded filament with the bound RecA enzyme then binds to a homologous DNA duplex, initiating DNA strand exchange and recombination. [6] Since the D. radiodurans genome lacks recB and recC genes, the identities of the helicase(s) and nuclease(s) that process broken DNA ends are not clear. There is, however, a gene encoding a protein called RecD2, which is similar to the RecD protein found in E. coli and other bacteria. [3, 6]
RecD2 from Deinococcus radiodurans belongs to the SF1 family of DNA helicases and unwinds DNA in the 5’-3’ direction using the energy from ATP hydrolysis [4]. Sequence analysis has shown that seven short amino acids motifs, which are conserved in SF1 helicases and found in E.coli RecD, are present in RecD2 as well. Yet compared to RecD found in E. coli, RecD2 is larger with an N-terminal extension of approximately 360-380 residues. This N-terminal region is not conserved within the RecD family members and do not contain sequences found in other helicases, indicating that RecD2 may have other functions. [1, 3, 6]
ATPase activity and DNA translocation
In the absence of DNA, RecD2 has no detectable ATPase activity, but in the presence of single-stranded DNA oligomers, RecD2 exhibits efficient ATP hydrolysis activity [1]. The rate-limiting step appears to be constituted by a structural rearrangement linked to Mg2+ coordination within the enzyme, which occurs before the hydrolysis step, according to a study from Toseland et al. In the absence of magnesium ions, hydrolysis is very slow. This process, which shuttles the enzyme structure between two conformations, appears to induce DNA movement and thus accounts for the tight coupling between translocation and ATPase activity. This mechanism has also been shown for the PcrA helicase and so may be a general mechanism for SF1 helicases. [4]
Helicase activity
RecD2 is able to unwind short DNA duplexes around 20 base pairs with either a 5’-single-stranded tail or a forked end, but not blunt-ended or 3’-tailed duplexes. Longer substrates are unwound much less efficiently. The enzyme acts catalytically, meaning that a single enzyme unwinds several DNA molecules. Duplexes with tails consisting of 10-12 nucleotides are bound tightly by RecD2, while DNA with shorter tails appear to be poor unwinding substrates since they are bound much less tightly. These results indicate that the enzyme requires very short DNA duplexes with at least a 10 nucleotide long 5’single-stranded tail for tight binding and efficient unwinding. [1]
One possible explanation for the inefficient unwinding of longer substrates is that RecD2 is a helicase with very low processivity, such that the enzyme is able to unwind shorter duplexes, but falls off before it can completely unwind longer substrates [1].This hypothesis was confirmed in a study by Shadrick et al., which demonstrated that RecD2 does have very low processivity [3]. A second possibility is that the individual DNA strands might rewind behind the enzyme as it travels along the DNA. A way to prevent rewinding in vitro is to include a single-stranded DNA-binding protein (SSB) in the mixture. In a study by Wang et al., the rate and extent of unwinding by RecD2 increased significantly in the presence of SSB from D. radiodurans. However, SSB from E. coli appeared to have little effect on the unwinding rate. [1]
DNA binding
The enzyme binds to 8 nucleotides of ssDNA in a cleft in the protein. This site contains a tyrosine residue supposedly acting as a pin to separate the strands during DNA unwinding. [3] A study conducted by Saikrishnan et al. showed that a pin-less mutant of RecD2 is a fully active ssDNA-dependent ATPase, but completely lacks helicase activity [2].
Effects of recD mutations
In a study by Servinsky et al., recD mutants were sensitive to ionizing radiation, UV light and hydrogen peroxide, but resistant to mitomycin C (MMC) and methyl methanesulfonate (MMS) [6]. These results indicate that RecD2 serves some function in DNA damage repair and/or recombination. The fact that the recD mutant is sensitive to ionizing radiation and hydrogen peroxide but not to MMC suggests that RecD2 might be involved in resistance to oxidation damage, something that has also been suggested by Zhou et al. [5, 6]
References:
[1] Wang, J., & Julin, D. A. (2004). DNA helicase activity of the RecD protein from Deinococcus radiodurans. Journal of Biological Chemistry, 279(50), 52024-52032.
[2] Saikrishnan, K., Griffiths, S. P., Cook, N., & Wigley, D. B. (2008). DNA binding to RecD: role of the 1B domain in SF1B helicase activity. The EMBO journal, 27(16), 2222-2229.
[3] Shadrick, W. R., & Julin, D. A. (2010). Kinetics of DNA unwinding by the RecD2 helicase from Deinococcus radiodurans. Journal of Biological Chemistry, 285(23), 17292-17300.
[4] Toseland, C. P., & Webb, M. R. (2013). ATPase Mechanism of the 5′-3′ DNA Helicase, RecD2 - Evidence for a Pre-hydrolysis Conformation Change. Journal of Biological Chemistry, 288(35), 25183-25193.
[5] Zhou, Q., Zhang, X., Xu, H., Xu, B., & Hua, Y. (2007). A new role of Deinococcus radiodurans RecD in antioxidant pathway. FEMS microbiology letters, 271(1), 118-125.
[6] Servinsky, M. D., & Julin, D. A. (2007). Effect of a recD mutation on DNA damage resistance and transformation in Deinococcus radiodurans. Journal of bacteriology, 189(14), 5101-5107.
PprA
It is believed that many different repair processes are active upon DNA damage in D. radiodurans. Non-homologous end-joining is thought to be one of these mechanisms, where PprA plays a critical role [2]. PprA is a pleiotropic protein promoting DNA repair in D. radiodurans. It acts by stimulating other enzymes such as ATP-dependent and NAD-dependent DNA ligases in the DNA repair process. [1, 2] Thermus thermophiles HB27 is the most closely related genus to Deinococcus, but does not contain a pprA homologue, emphasizing the uniqueness of the DNA repair mechanisms of D. radiodurans [2].
DNA binding
PprA is able to form complexes with either dsDNA in an open circular form or linear dsDNA, but not dsDNA in a closed circular form or ssDNA, indicating that it binds preferentially to dsDNA containing strand breaks. PprA is also able to inhibit the activity of exonucleases when bound to DNA, thus protecting the strands from degradation. [2]
Stimulation of enzymes
PprA has been shown to be able to stimulate DNA ligation catalyzed by T4 DNA ligase and E. coli DNA ligase, although PprA itself does not possess DNA ligation activity. Narumi et al. showed that the degree of stimulation was dependent on the concentration of PprA. [2] Upon being phosphorylated, PprA increases its DNA binding affinity for dsDNA and is thereby able to stimulate T4 DNA ligase at a much higher rate. [4] In another study by Kota et al., PprA was also able to stimulate the activity of catalase from E. coli both in vitro and in vivo. The transgenic E. coli cells expressing PprA has been shown to have a higher tolerance to hydrogen peroxide as compared to wild type cells due to an increase in antioxidant enzyme activity. [3]
References:
[1] Tanaka, M., Earl, A. M., Howell, H. A., Park, M. J., Eisen, J. A., Peterson, S. N., & Battista, J. R. (2004). Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics, 168(1), 21-33.
[2] Narumi, I., Satoh, K., Cui, S., Funayama, T., Kitayama, S., & Watanabe, H. (2004). PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Molecular microbiology, 54(1), 278-285.
[3] Kota, S., & Misra, H. S. (2006). PprA: A protein implicated in radioresistance of Deinococcus radiodurans stimulates catalase activity in Escherichia coli. Applied microbiology and biotechnology, 72(4), 790-796.
[4] Rajpurohit, Y. S., & Misra, H. S. (2013). Structure-function study of deinococcal serine/threonine protein kinase implicates its kinase activity and DNA repair protein phosphorylation roles in radioresistance of Deinococcus radiodurans. The international journal of biochemistry & cell biology, 45(11), 2541-2552.
Safety Switch
It is our view as a team, that taking responsibility for the work conducted is of highest importance. One of the main aspects of this commitment is the prevention of modified organisms ever escaping the laboratory and accidentally ending up in nature. For this purpose, a genetic construct has been designed called the safety switch, which will serve to address both ethical and safety issues regarding the manufacturing of a more resilient yeast strain.
The SUC2 regulation in Saccharomyces cerevisiae
The SUC2 gene in Saccharomyces cerevisiae codes for the enzyme invertase, which catalyses the hydrolysation of the glycosidic bond in the sucrose dimer, breaking it down into its constituent monomers, glucose and fructose[1]. This enzyme is important to the survivability of the yeast cells since it allows them to utilise sucrose as an alternative carbon source should they ever be in a glucose deprived environment. However, this is a less efficient way of obtaining energy than simply metabolising glucose, hence it should be used exclusively when glucose is in short demand. Thus, in order to regulate this, the expression of the SUC2 gene is sensitive to the amount of ATP available to the cell[2].
This ATP sensitivity originates from the repression of the SUC2 gene, when enough ATP is present, using a repressor complex which is made up by the repressor protein MIG1 and the general repressor complex SSN6-TUP1. This conjoined repressor binds to the upstream promoter region of the SUC2 gene[3], where it will prevent transcription. However, the assembly of this repressor complex can be hindered by the protein kinase SNF1. A SNF1-dependent phosphorylation of the MIG1 protein makes it unable to interact with the SSN6-TUP1 complex[4], resulting in the SUC2 gene being expressed. Furthermore, the SNF1 kinase will, in turn, be blocked by the presence of glucose, resulting in a glucose dependent mechanism for the expression of the SUC2 gene[2].
This means that when glucose is present in high enough concentration, the SUC2 gene will remain repressed due to the formation of the repressor complex. Thus, when the glucose concentration is low, the repressor complex cannot assemble which results in the gene being expressed.
The Safety Switch
Our ambition was to design a genetic construct with the ability to sense whether the cells were still in the lab, or not. For this purpose, we chose the glucose dependence of the SUC2 gene as a way for the cells to sense their environment. This was mainly because we can ensure that the glucose levels are always kept high when working with the cells in the lab, as well as we know that the access to glucose will be much more scarce at any other location. This makes the SUC2 regulation a very appropriate choice for making the cells “understand” when they are no longer in the right place.
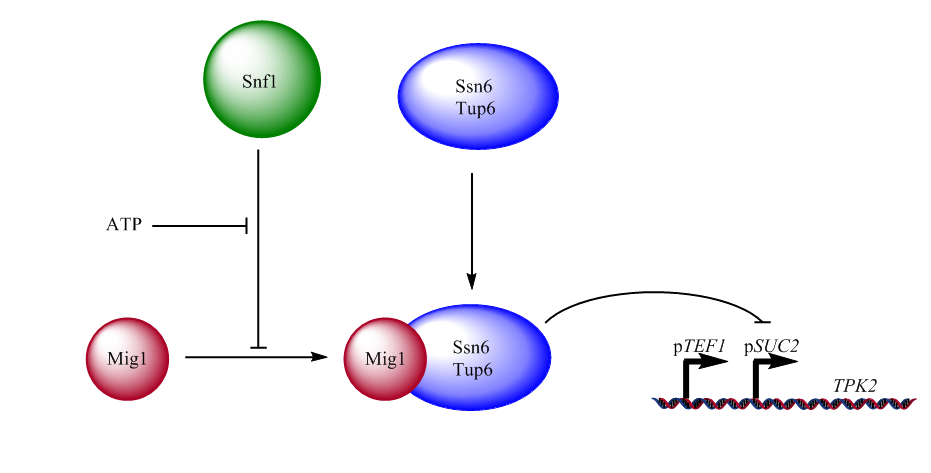
The Safety Switch utilises this system of regulation, except for the gene itself. The SUC2 gene has been replaced with a new gene, which is harmful to the cells, and which will be expressed instead when the glucose concentration is low. For this purpose, we have chosen the gene TPK2 which only becomes dangerous to the cells when it is overexpressed[6]. This choice was motivated by the fact that the repressor of the gene is not perfect, and there will always be small expression of the gene, even when repressed. The fact that a certain amount of TPK2 is required before it becomes dangerous to the cells makes sure that they are not accidentally harmed due to the marginal inefficiency of the repressor. However, this also poses a potential problem when the Safety Switch is activated, since the expression needs to be sufficiently strong for the TPK2 to have the desired effect. Therefore, in addition to the pSUC2 promoter, another promoter called pTEF1 is added upstream of the pSUC2. This is because we want to elevate the expression level of the gene, without altering the way it is regulated. Due to the fact that the pSUC2 region lies in between the new promoter and the gene, the pTEF1 promoter cannot activate the expression of the gene as long as the repressor is bound to the pSUC2. Furthermore, the short length of the pSUC2 promoter allows the pTEF1 promoter to activate expression of TPK2, even though they are not adjacent to each other.
Expression of mRFP (monomeric Red Fluorescent Protein) was used to analyze expression levels of the safety switch. To evaluate the effect of connecting pTEF1 to pSUC2, two more versions of the safety switch were made: pTEF1-pSUC2-mRFP and pSUC2-mRFP. This allowed comparison of gene expression between these two set ups in order to determine whether or not the addition of pTEF1 actually resulted in a higher level of gene expression.
References:
[1] Saccharomyces Genome Database (2010) SUC2 / YIL162W http://www.yeastgenome.org/locus/SUC2/overview (September 17:th 2015)
[2] García‐Salcedo, R., Lubitz, T., Beltran, G., Elbing, K., Tian, Y., Frey, S., ... & Hohmann, S. (2014). Glucose de‐repression by yeast AMP‐activated protein kinase SNF1 is controlled via at least two independent steps. Febs Journal, 281(7), 1901-1917.(September 17:th 2015)
[3] Özcan, S., Vallier, L. G., Flick, J. S., Carlson, M., & Johnston, M. (1997). Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast, 13(2), 127-137.(September 17:th 2015)
[4] Papamichos‐Chronakis, M., Gligoris, T., & Tzamarias, D. (2004). The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8‐Tup1 co‐repressor. EMBO reports, 5(4), 368-372.(September 17:th 2015)
[5] Saccharomyces Genome Database (2007) TPK2 / YPL203W http://www.yeastgenome.org/locus/Tpk2/overview (September 17:th 2015)