Difference between revisions of "Team:DTU-Denmark/Project/POC MAGE"
| Line 60: | Line 60: | ||
> | > | ||
| − | <a href=" | + | <a href="" |
class="dropdown-toggle" | class="dropdown-toggle" | ||
data-toggle="dropdown" | data-toggle="dropdown" | ||
| Line 76: | Line 76: | ||
<li | <li | ||
| + | |||
| + | |||
| Line 97: | Line 99: | ||
> | > | ||
| − | <a href=" | + | <a href="" |
class="dropdown-toggle" | class="dropdown-toggle" | ||
data-toggle="dropdown" | data-toggle="dropdown" | ||
role="button" | role="button" | ||
| − | aria-expanded="false">Project | + | aria-expanded="false">Project description |
<span class="caret"></span></a><ul class="dropdown-menu" role="menu"> | <span class="caret"></span></a><ul class="dropdown-menu" role="menu"> | ||
| + | <li > | ||
| + | <a href="/Team:DTU-Denmark/Project" | ||
| + | >Pernille og Scott | ||
| + | </a></li> | ||
<li > | <li > | ||
<a href="/Team:DTU-Denmark/Project/Overview" | <a href="/Team:DTU-Denmark/Project/Overview" | ||
| Line 137: | Line 143: | ||
<li class="active"> | <li class="active"> | ||
<a href="/Team:DTU-Denmark/Project/POC_MAGE" | <a href="/Team:DTU-Denmark/Project/POC_MAGE" | ||
| − | > | + | >Proof of concept of MAGE in B. subtilis |
</a></li></ul></li> | </a></li></ul></li> | ||
<li > | <li > | ||
| Line 143: | Line 149: | ||
>Human Practices | >Human Practices | ||
</a></li> | </a></li> | ||
| + | <li | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | > | ||
| + | <a href="" | ||
| + | class="dropdown-toggle" | ||
| + | data-toggle="dropdown" | ||
| + | role="button" | ||
| + | aria-expanded="false">Parts | ||
| + | <span class="caret"></span></a><ul class="dropdown-menu" role="menu"> | ||
<li > | <li > | ||
| − | <a href="/Team:DTU-Denmark/ | + | <a href="/Team:DTU-Denmark/Parts/Parts" |
>Parts | >Parts | ||
</a></li> | </a></li> | ||
| + | <li > | ||
| + | <a href="/Team:DTU-Denmark/Description" | ||
| + | >Characterisation of xylR | ||
| + | </a></li></ul></li> | ||
<li > | <li > | ||
<a href="/Team:DTU-Denmark/Journal" | <a href="/Team:DTU-Denmark/Journal" | ||
| Line 164: | Line 187: | ||
> | > | ||
| − | <a href=" | + | <a href="" |
class="dropdown-toggle" | class="dropdown-toggle" | ||
data-toggle="dropdown" | data-toggle="dropdown" | ||
| Line 206: | Line 229: | ||
<div class="container-fluid" id="scrollspy"> | <div class="container-fluid" id="scrollspy"> | ||
<ul class="nav navbar-nav"> | <ul class="nav navbar-nav"> | ||
| − | <li><a class="page-scroll" href="#Overview">Overview</a></li><li><a class="page-scroll" href="#Methods">Methods</a></li> | + | <li><a class="page-scroll" href="#Overview">Overview</a></li><li><a class="page-scroll" href="#Achievements">Achievements</a></li><li><a class="page-scroll" href="#MAGE-competent-strains">MAGE competent strains</a></li><li><a class="page-scroll" href="#Methods-for-proof-of-concept">Methods for proof of concept</a></li><li><a class="page-scroll" href="#Discussion">Discussion</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
| Line 221: | Line 244: | ||
<h3><br></h3> | <h3><br></h3> | ||
| − | <h1> | + | <h1>Proof of concept of MAGE in B. subtilis</h1> |
<hr/> | <hr/> | ||
| Line 231: | Line 254: | ||
<li> | <li> | ||
| − | <a id="Methods-submenu" class="btn btn-default btn-transparent btn-lg page-scroll" href="#Methods">Methods</a> | + | <a id="Achievements-submenu" class="btn btn-default btn-transparent btn-lg page-scroll" href="#Achievements">Achievements</a> |
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a id="MAGE-competent-strains-submenu" class="btn btn-default btn-transparent btn-lg page-scroll" href="#MAGE-competent-strains">MAGE competent strains</a> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a id="Methods-for-proof-of-concept-submenu" class="btn btn-default btn-transparent btn-lg page-scroll" href="#Methods-for-proof-of-concept">Methods for proof of concept</a> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a id="Discussion-submenu" class="btn btn-default btn-transparent btn-lg page-scroll" href="#Discussion">Discussion</a> | ||
</li> | </li> | ||
| Line 242: | Line 277: | ||
<li> | <li> | ||
| − | <a id="Methods-submenu" class="page-scroll" href="#Methods">Methods</a> | + | <a id="Achievements-submenu" class="page-scroll" href="#Achievements">Achievements</a> |
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a id="MAGE-competent-strains-submenu" class="page-scroll" href="#MAGE-competent-strains">MAGE competent strains</a> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a id="Methods-for-proof-of-concept-submenu" class="page-scroll" href="#Methods-for-proof-of-concept">Methods for proof of concept</a> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a id="Discussion-submenu" class="page-scroll" href="#Discussion">Discussion</a> | ||
</li> | </li> | ||
| Line 266: | Line 313: | ||
Overview | Overview | ||
</h1> | </h1> | ||
| − | <p>In order for MAGE (Multiplex Automated Genome Engineering) to work at a high efficiency, a strain with inserted recombinase and inhibited or knocked out mismatch repair gene has to be used [1]. In our project, two different recombinases were used: a recombination protein Beta from the E. coli phage Lambda, which was codon optimized for B. subtilis 168, and GP35, a recombinase from the B.subtilis phage SPP1 [2]. The mismatch repair proteins known as MutS and MutL were knocked out by transforming pSB1C3_recombinase plasmid into the B. subtilis W168. Since the MutL protein is dependent on the binding of MutS, the knockout of the mutS disables the function of the MutL protein [3]. | + | <p>In order for MAGE (Multiplex Automated Genome Engineering) to work at a high efficiency, a strain with inserted recombinase and inhibited or knocked out mismatch repair gene has to be used [1]. In our project, two different recombinases were used: a recombination protein Beta from the <em>E. coli</em> phage Lambda, which was codon optimized for B. subtilis 168, and GP35, a recombinase from the B.subtilis phage SPP1 [2]. The mismatch repair proteins known as MutS and MutL were knocked out by transforming pSB1C3_recombinase plasmid into the B. subtilis W168. Since the MutL protein is dependent on the binding of MutS, the knockout of the mutS disables the function of the MutL protein [3].<br /> |
| + | Four <em>Bacillus subtilis</em> strains which expressed a recombinase were created by genetically engineering the wild type strain 168:</p> | ||
<ul> | <ul> | ||
| Line 274: | Line 322: | ||
<li><em>∆mutS::GP35-neoR</em></li> | <li><em>∆mutS::GP35-neoR</em></li> | ||
</ul> | </ul> | ||
| + | |||
| + | <p>For proof of concept we decided to make a single protein substitution in the ribosomal S12 protein in B. subtilis results in resistance to the antibiotic streptomycin. The required change is a lysine to arginine substitution at position 56 of the protein. The S12 subunit is coded by the rpsL gene [4]. An oligo that could integrate this change was design using MODEST [5]. This oligos were successfully integrated into the two strains: <em>∆mutS::beta-neo<sup>R</sup></em> and <em>∆mutS::GP35-neo<sup>R</sup>.</em></p> | ||
</div> | </div> | ||
| Line 280: | Line 330: | ||
| − | <div id=" | + | <div id="Achievements"> |
<div class="container"> | <div class="container"> | ||
<div class="row col-md-12"> | <div class="row col-md-12"> | ||
<h1> | <h1> | ||
| − | + | Achievements | |
| + | </h1> | ||
| + | <ul> | ||
| + | <li>Made <em>Bacillus subtilis 168</em> WT MAGE compatible</li> | ||
| + | <li>Showed that the MODEST program can make oligoes that can be inserted in <em>Bacillus. subtilis 168.</em></li> | ||
| + | <li>Introducing Multiplex Automated Genome Engineering (MAGE) in <em>Bacillus subtilis 168</em>.</li> | ||
| + | </ul> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div id="MAGE-competent-strains"> | ||
| + | <div class="container"> | ||
| + | <div class="row col-md-12"> | ||
| + | <h1> | ||
| + | MAGE competent strains | ||
</h1> | </h1> | ||
<p>All the strain were made by homologous recombineering. Plasmids containing cassettes that were able to do a double-crossover homologous recombineering into the genome of B. subtilis 168 (referred to as knockout (KO) plasmid). These were used to, simultaneously, delete the desired gene (amyE or mutS) and inserting the expression cassette for one of the recombinases: beta or GP35.</p> | <p>All the strain were made by homologous recombineering. Plasmids containing cassettes that were able to do a double-crossover homologous recombineering into the genome of B. subtilis 168 (referred to as knockout (KO) plasmid). These were used to, simultaneously, delete the desired gene (amyE or mutS) and inserting the expression cassette for one of the recombinases: beta or GP35.</p> | ||
<p><img alt="" src="/wiki/images/a/a4/DTU-Denmark_pDG268neo_recombinase.png" style="width: 400px; height: 397px;" /><img alt="" src="/wiki/images/c/c1/DTU-Denmark_pSB1C3neo_recombinase.png" style="width: 400px; height: 397px;" /></p> | <p><img alt="" src="/wiki/images/a/a4/DTU-Denmark_pDG268neo_recombinase.png" style="width: 400px; height: 397px;" /><img alt="" src="/wiki/images/c/c1/DTU-Denmark_pSB1C3neo_recombinase.png" style="width: 400px; height: 397px;" /></p> | ||
| + | |||
| + | <p><span style="font-size:14px;">Figur 1. Shows the general concepts of the two plasmids pDG268neo_recombinase and pSB1C3_recombinase. Both exists in two versions, one with each of the recombinase proteins CDSs (Beta and GP35). Those also have different RBSs since they are optimized for the CDS. Upstream of neoR is a promoter and RBS and downstream of neoR is a terminator, but sequences and positions of these features are not known.</span></p> | ||
| + | |||
| + | <p> </p> | ||
<p>Four different plasmids was assembled to make the four MAGE ready strains:<br /> | <p>Four different plasmids was assembled to make the four MAGE ready strains:<br /> | ||
| Line 295: | Line 366: | ||
pSB1C3_Beta-neoR<br /> | pSB1C3_Beta-neoR<br /> | ||
pSB1C3_GP35-neoR</p> | pSB1C3_GP35-neoR</p> | ||
| − | |||
| − | |||
| − | |||
<p> </p> | <p> </p> | ||
| Line 329: | Line 397: | ||
<p><img alt="" src="/wiki/images/d/d9/DTU-Denmark_pDG268_1.png" style="width: 700px; height: 835px;" /></p> | <p><img alt="" src="/wiki/images/d/d9/DTU-Denmark_pDG268_1.png" style="width: 700px; height: 835px;" /></p> | ||
| − | <p> </p> | + | <p><span style="font-size:14px;">Figur 2. Gibsom assembly of the pDG268neo_GP35, the gblocks was fused to the “gp35 Gblocks fused” in the same reaction. The Gibsom assembly of pDG268neo_beta was similar.</span></p> |
| − | <p> | + | <p> </p> |
<p>This resulted in two different plasmids pDG268neo_beta and pDG268neo_gp35.</p> | <p>This resulted in two different plasmids pDG268neo_beta and pDG268neo_gp35.</p> | ||
| Line 364: | Line 432: | ||
<p><img alt="" src="/wiki/images/d/dc/DTU-Denmark_pDG268_2.png" style="width: 1413px; height: 720px;" /></p> | <p><img alt="" src="/wiki/images/d/dc/DTU-Denmark_pDG268_2.png" style="width: 1413px; height: 720px;" /></p> | ||
| − | <p>< | + | <p><span style="font-size:14px;">Figur 3. Gibsom assembly of the pSB1C3_beta. The feature called “mutL” corresponding mutS downstream. The Gibsom assembly of pSB1C3_GP35 was similar.</span></p> |
<p>All plasmids were verified by restriction enzyme digestion.</p> | <p>All plasmids were verified by restriction enzyme digestion.</p> | ||
| Line 372: | Line 440: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div id="Methods-for-proof-of-concept"> | ||
| + | <div class="container"> | ||
| + | <div class="row col-md-12"> | ||
| + | <h1> | ||
| + | Methods for proof of concept | ||
| + | </h1> | ||
| + | <p>Electroporation competent cells were prepared in following the protocol: Electroporation competent <em>Bacillus subtilis 168 (missing link). </em>Electropration was carried out according to the MAGE in <em>Bacillus subtilis 168</em> protocol. The colonies from the LB plates were colonies picked onto an LB and LB + 500y streptomycin (strep) plates. Plates were incubated at 37 degC for 48 hours and CFUs were counted on both the LB and the strep plates.Results</p> | ||
| + | |||
| + | <div aria-multiselectable="true" class="panel-group" id="accordion" role="tablist"> | ||
| + | <div class="panel panel-default"> | ||
| + | <div class="panel-heading" id="headingOne" role="tab"> | ||
| + | <h4 class="panel-title"><a aria-controls="collapseOne" aria-expanded="false" class="collapsed" data-parent="#accordion" data-toggle="collapse" href="#collapseOne" role="button">Results</a></h4> | ||
| + | </div> | ||
| + | |||
| + | <div aria-labelledby="headingOne" class="panel-collapse collapse" id="collapseOne" role="tabpanel"> | ||
| + | <div class="panel-body"> | ||
| + | <div class="panel-body"> | ||
| + | <table border="1" cellpadding="1" cellspacing="1" class="table table" style="width:500px;"> | ||
| + | <thead> | ||
| + | <tr> | ||
| + | <th scope="col"> </th> | ||
| + | <th scope="col" style="text-align: center;">Total number of CFUs</th> | ||
| + | <th scope="col" style="text-align: center;">Number of transformant</th> | ||
| + | <th scope="col" style="text-align: center;">Transformation frequency</th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td>Δ<i>mutS::beta-neo</i><i><sup>R</sup></i></td> | ||
| + | <td style="text-align: center;">52</td> | ||
| + | <td style="text-align: center;">7</td> | ||
| + | <td style="text-align: center;">0.13</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Δ<i>mutS::GP35-neo</i><i><sup>R</sup></i></td> | ||
| + | <td style="text-align: center;">100</td> | ||
| + | <td style="text-align: center;">1</td> | ||
| + | <td style="text-align: center;">0.01</td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | |||
| + | <p><span style="font-size:14px;">Table 1. shows the data for the MAGE proof of concept.</span></p> | ||
| + | |||
| + | <p> </p> | ||
| + | |||
| + | <p>Second replication of the plates:</p> | ||
| + | |||
| + | <p><img alt="" src="\files\POC mutSgp35.png" style="width: 375px; height: 205px;" /><img alt="" src="\files\POC mutSbeta.png" style="width: 397px; height: 205px;" /></p> | ||
| + | |||
| + | <p><span style="font-size:14px;">Figur 4. Pictures shows the first and second replication of the transformants. From left to right of ΔmutS::GP35-neo<sup>R</sup> <span style="font-size:14px;"> replicate1</span>, <em>ΔmutS::GP35-neo</em><sup><em>R</em> </sup><span style="font-size:14px;">replicate2 </span>, Δ<i>mutS::beta-neo</i><i><sup>R </sup></i>replica1, and <span style="font-size:14px;">Δ<i>mutS::beta-neo</i><i><sup>R </sup></i>replica2.</span></span></p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div id="Discussion"> | ||
| + | <div class="container"> | ||
| + | <div class="row col-md-12"> | ||
| + | <h1> | ||
| + | Discussion | ||
| + | </h1> | ||
| + | <p>The results of this proof of concept indicates that MAGE works in two of the MAGE compatible strains Δ<em>mutS::beta-neo</em><em><sup>R</sup></em> and Δ<em>mutS::GP35-neo</em><em><sup>R</sup></em><em>. </em>More thoroughly experiments needs to be done. The result suggest that the beta protein is a more effective recombinase for the type of oligo used in this experiment.</p> | ||
</div> | </div> | ||
Revision as of 03:09, 19 September 2015
Proof of concept of MAGE in B. subtilis
Overview
In order for MAGE (Multiplex Automated Genome Engineering) to work at a high efficiency, a strain with inserted recombinase and inhibited or knocked out mismatch repair gene has to be used [1]. In our project, two different recombinases were used: a recombination protein Beta from the E. coli phage Lambda, which was codon optimized for B. subtilis 168, and GP35, a recombinase from the B.subtilis phage SPP1 [2]. The mismatch repair proteins known as MutS and MutL were knocked out by transforming pSB1C3_recombinase plasmid into the B. subtilis W168. Since the MutL protein is dependent on the binding of MutS, the knockout of the mutS disables the function of the MutL protein [3].
Four Bacillus subtilis strains which expressed a recombinase were created by genetically engineering the wild type strain 168:
- ∆amyE::beta-neoR
- ∆amyE::GP35-neoR
- ∆mutS::beta-neoR
- ∆mutS::GP35-neoR
For proof of concept we decided to make a single protein substitution in the ribosomal S12 protein in B. subtilis results in resistance to the antibiotic streptomycin. The required change is a lysine to arginine substitution at position 56 of the protein. The S12 subunit is coded by the rpsL gene [4]. An oligo that could integrate this change was design using MODEST [5]. This oligos were successfully integrated into the two strains: ∆mutS::beta-neoR and ∆mutS::GP35-neoR.
Achievements
- Made Bacillus subtilis 168 WT MAGE compatible
- Showed that the MODEST program can make oligoes that can be inserted in Bacillus. subtilis 168.
- Introducing Multiplex Automated Genome Engineering (MAGE) in Bacillus subtilis 168.
MAGE competent strains
All the strain were made by homologous recombineering. Plasmids containing cassettes that were able to do a double-crossover homologous recombineering into the genome of B. subtilis 168 (referred to as knockout (KO) plasmid). These were used to, simultaneously, delete the desired gene (amyE or mutS) and inserting the expression cassette for one of the recombinases: beta or GP35.
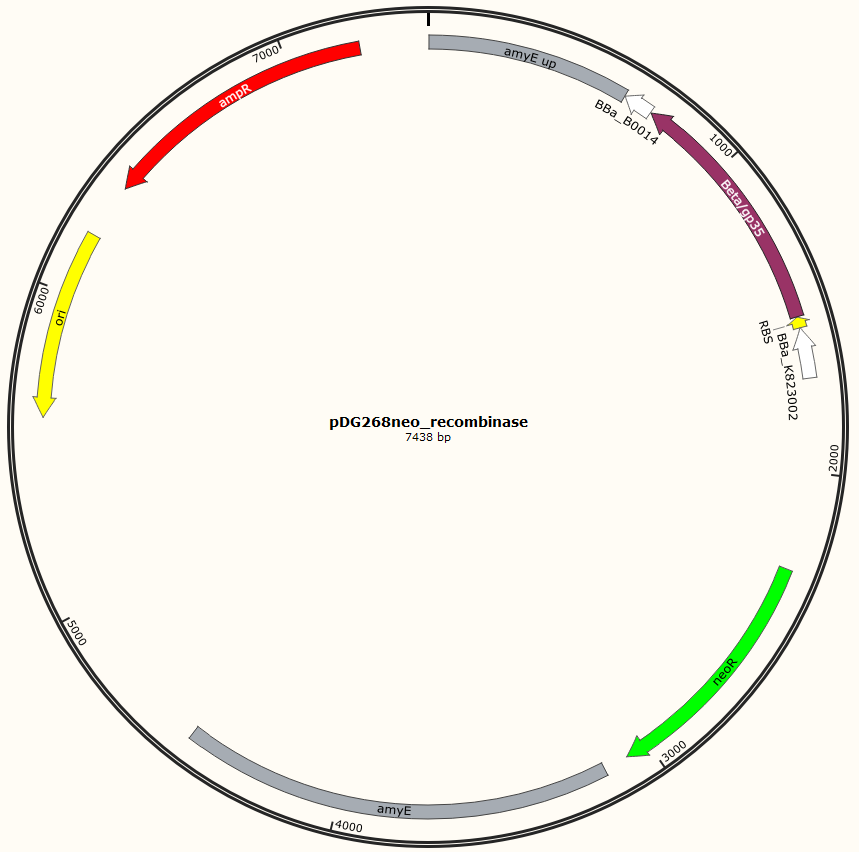
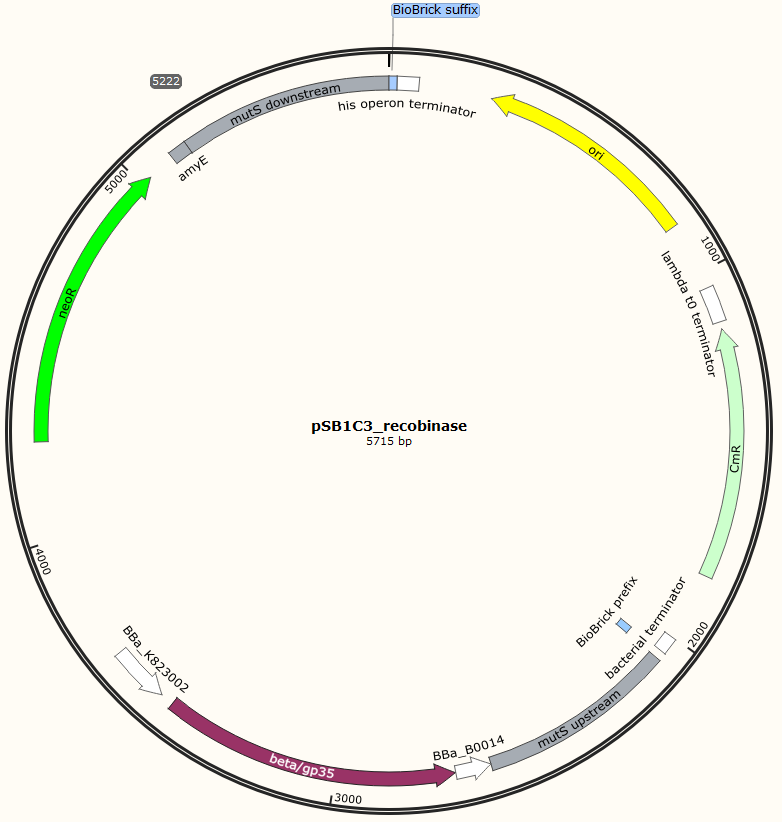
Figur 1. Shows the general concepts of the two plasmids pDG268neo_recombinase and pSB1C3_recombinase. Both exists in two versions, one with each of the recombinase proteins CDSs (Beta and GP35). Those also have different RBSs since they are optimized for the CDS. Upstream of neoR is a promoter and RBS and downstream of neoR is a terminator, but sequences and positions of these features are not known.
Four different plasmids was assembled to make the four MAGE ready strains:
pDG268neo_Beta-neoR
pDG268neo_GP35-neoR
pSB1C3_Beta-neoR
pSB1C3_GP35-neoR
For pDG268neo_recobinase a DNA sequence containing following features
Missing: make a table on following
● Promoter: PliaG from BBa_K823002 was used.
● RBSs were optimized for the specific CDS using the salis lab RBS calculator (Missing https://www.denovodna.com/software/)
● CDS for recombination protein beta or GP35
● Terminator: we use rho-independent Part:BBa_B0014
Sequence was ordered from IDT as two gblocks (for each recombinase) with overlapping regions, thus they can be assembled with Gibsom assembly.
pDG268neo was linearized in a PCR with primers, so that the native lacZ was omitted. This linearized plasmid was purified and used in a Gibsom assembly reaction with two cognate gblocks. This is shown on the figure missing.
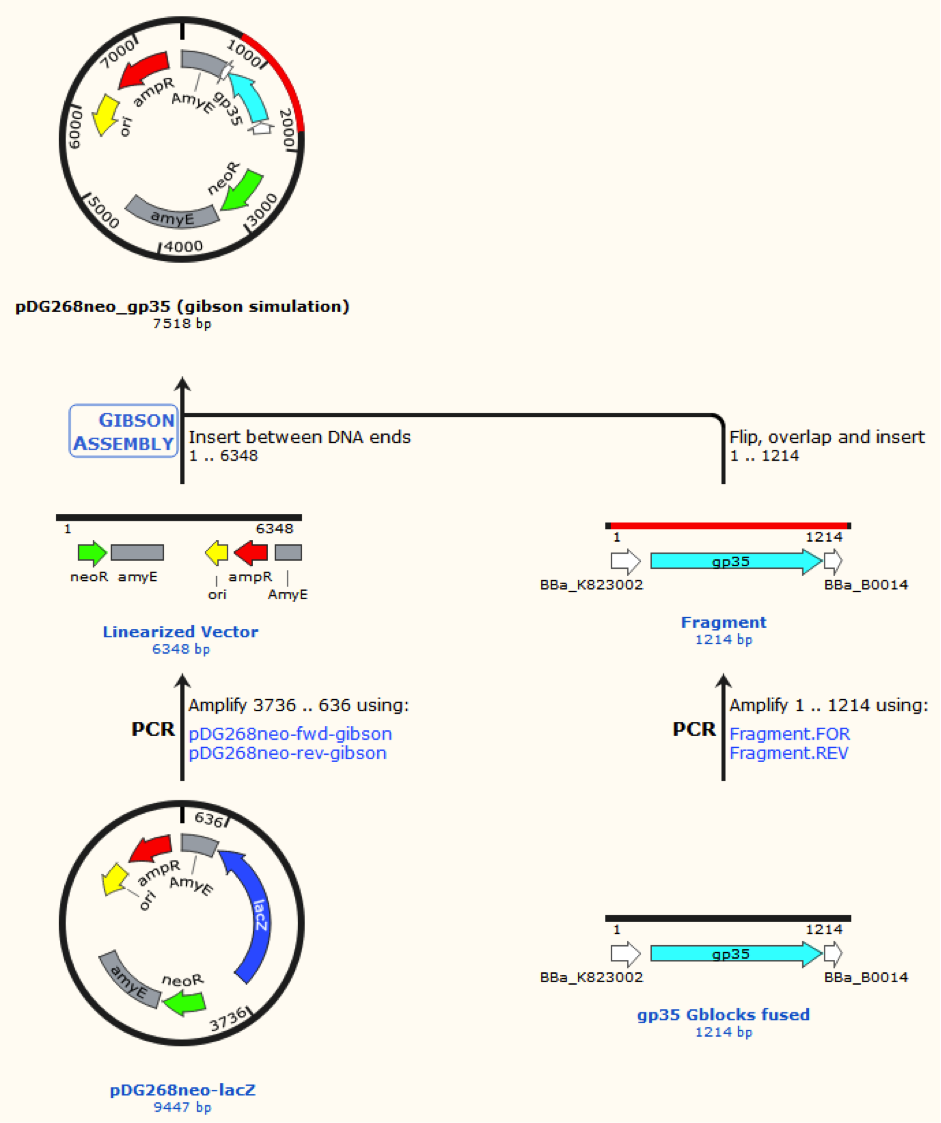
Figur 2. Gibsom assembly of the pDG268neo_GP35, the gblocks was fused to the “gp35 Gblocks fused” in the same reaction. The Gibsom assembly of pDG268neo_beta was similar.
This resulted in two different plasmids pDG268neo_beta and pDG268neo_gp35.
The two mutS KO plasmids was made of the following DNA fragments:
1. About 500 bp up- and downstream from mutS was amplified, using primers with tails.
2. Recombinase and neoR expression cassettes was amplified from the pDG268neo_recombinase
3. Linearized pSB1C3 was used as template.
These fragments was assembled into two different plasmids pSB1C3_mutS::beta-neoR and pSB1C3_mutS::gp35-neoR using Gibsom assembly.
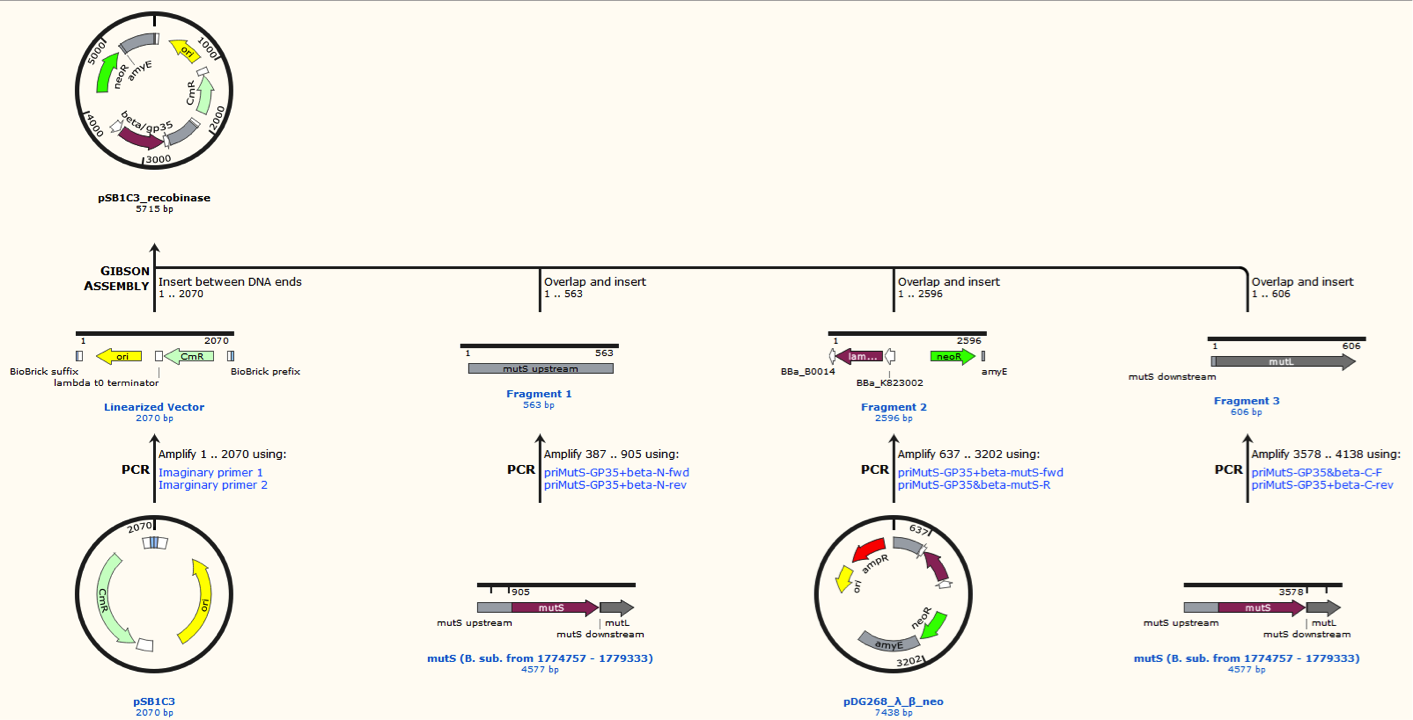
Figur 3. Gibsom assembly of the pSB1C3_beta. The feature called “mutL” corresponding mutS downstream. The Gibsom assembly of pSB1C3_GP35 was similar.
All plasmids were verified by restriction enzyme digestion.
We prepared naturel competent B. subtilis 168 to be transformed. The plasmids were linearized with restriction enzymes. Then transformed into naturally competent B. subtilis 168, transformants were selected on 5ɣ neo. Transformants were verified with colony PCRs.
Methods for proof of concept
Electroporation competent cells were prepared in following the protocol: Electroporation competent Bacillus subtilis 168 (missing link). Electropration was carried out according to the MAGE in Bacillus subtilis 168 protocol. The colonies from the LB plates were colonies picked onto an LB and LB + 500y streptomycin (strep) plates. Plates were incubated at 37 degC for 48 hours and CFUs were counted on both the LB and the strep plates.Results
| Total number of CFUs | Number of transformant | Transformation frequency | |
|---|---|---|---|
| ΔmutS::beta-neoR | 52 | 7 | 0.13 |
| ΔmutS::GP35-neoR | 100 | 1 | 0.01 |
Table 1. shows the data for the MAGE proof of concept.
Second replication of the plates:


Figur 4. Pictures shows the first and second replication of the transformants. From left to right of ΔmutS::GP35-neoR replicate1, ΔmutS::GP35-neoR replicate2 , ΔmutS::beta-neoR replica1, and ΔmutS::beta-neoR replica2.
Discussion
The results of this proof of concept indicates that MAGE works in two of the MAGE compatible strains ΔmutS::beta-neoR and ΔmutS::GP35-neoR. More thoroughly experiments needs to be done. The result suggest that the beta protein is a more effective recombinase for the type of oligo used in this experiment.
Department of Systems Biology
Søltofts Plads 221
2800 Kgs. Lyngby
Denmark
P: +45 45 25 25 25
M: dtu-igem-2015@googlegroups.com