Difference between revisions of "Team:DTU-Denmark/Project/POC MAGE"
(Created page with "<html> <head> <meta name="viewport" content="width=device-width, initial-scale=1, maximum-scale=1, user-scalable=no"> <meta http-equiv="X-UA-Compatible" content="IE=...") |
|||
| Line 206: | Line 206: | ||
<div class="container-fluid" id="scrollspy"> | <div class="container-fluid" id="scrollspy"> | ||
<ul class="nav navbar-nav"> | <ul class="nav navbar-nav"> | ||
| − | <li><a class="page-scroll" href="#Methods">Methods</a></li> | + | <li><a class="page-scroll" href="#Overview">Overview</a></li><li><a class="page-scroll" href="#Methods">Methods</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
| Line 225: | Line 225: | ||
<hr/> | <hr/> | ||
<ul class="list-inline hidden-xs hidden-sm"> | <ul class="list-inline hidden-xs hidden-sm"> | ||
| + | |||
| + | <li> | ||
| + | <a id="Overview-submenu" class="btn btn-default btn-transparent btn-lg page-scroll" href="#Overview">Overview</a> | ||
| + | </li> | ||
<li> | <li> | ||
| Line 232: | Line 236: | ||
</ul> | </ul> | ||
<ul class="list-inline visible-xs visible-sm"> | <ul class="list-inline visible-xs visible-sm"> | ||
| + | |||
| + | <li> | ||
| + | <a id="Overview-submenu" class="page-scroll" href="#Overview">Overview</a> | ||
| + | </li> | ||
<li> | <li> | ||
| Line 243: | Line 251: | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
Scroll down for more<br> | Scroll down for more<br> | ||
| − | <a href="# | + | <a href="#Overview" class="btn btn-circle btn-transparent page-scroll"> |
<i class="fa fa-angle-double-down"></i> | <i class="fa fa-angle-double-down"></i> | ||
</a> | </a> | ||
| Line 251: | Line 259: | ||
</div> | </div> | ||
| + | |||
| + | <div id="Overview"> | ||
| + | <div class="container"> | ||
| + | <div class="row col-md-12"> | ||
| + | <h1> | ||
| + | Overview | ||
| + | </h1> | ||
| + | <p>In order for MAGE (Multiplex Automated Genome Engineering) to work at a high efficiency, a strain with inserted recombinase and inhibited or knocked out mismatch repair gene has to be used [1]. In our project, two different recombinases were used: a recombination protein Beta from the E. coli phage Lambda, which was codon optimized for B. subtilis 168, and GP35, a recombinase from the B.subtilis phage SPP1 [2]. The mismatch repair proteins known as MutS and MutL were knocked out by transforming pSB1C3_recombinase plasmid into the B. subtilis W168. Since the MutL protein is dependent on the binding of MutS, the knockout of the mutS disables the function of the MutL protein [3]. For doing proof of concept of MAGE in B. subtilis 168 four different strains was produced via genetic recombineering. All derived from Bacillus subtilis 168 WT:</p> | ||
| + | |||
| + | <ul> | ||
| + | <li><em>∆amyE::beta-neoR</em></li> | ||
| + | <li><em>∆amyE::GP35-neoR</em></li> | ||
| + | <li><em>∆mutS::beta-neoR</em></li> | ||
| + | <li><em>∆mutS::GP35-neoR</em></li> | ||
| + | </ul> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
<div id="Methods"> | <div id="Methods"> | ||
Revision as of 00:03, 19 September 2015
Overview
In order for MAGE (Multiplex Automated Genome Engineering) to work at a high efficiency, a strain with inserted recombinase and inhibited or knocked out mismatch repair gene has to be used [1]. In our project, two different recombinases were used: a recombination protein Beta from the E. coli phage Lambda, which was codon optimized for B. subtilis 168, and GP35, a recombinase from the B.subtilis phage SPP1 [2]. The mismatch repair proteins known as MutS and MutL were knocked out by transforming pSB1C3_recombinase plasmid into the B. subtilis W168. Since the MutL protein is dependent on the binding of MutS, the knockout of the mutS disables the function of the MutL protein [3]. For doing proof of concept of MAGE in B. subtilis 168 four different strains was produced via genetic recombineering. All derived from Bacillus subtilis 168 WT:
- ∆amyE::beta-neoR
- ∆amyE::GP35-neoR
- ∆mutS::beta-neoR
- ∆mutS::GP35-neoR
Methods
All the strain were made by homologous recombineering. Plasmids containing cassettes that were able to do a double-crossover homologous recombineering into the genome of B. subtilis 168 (referred to as knockout (KO) plasmid). These were used to, simultaneously, delete the desired gene (amyE or mutS) and inserting the expression cassette for one of the recombinases: beta or GP35.
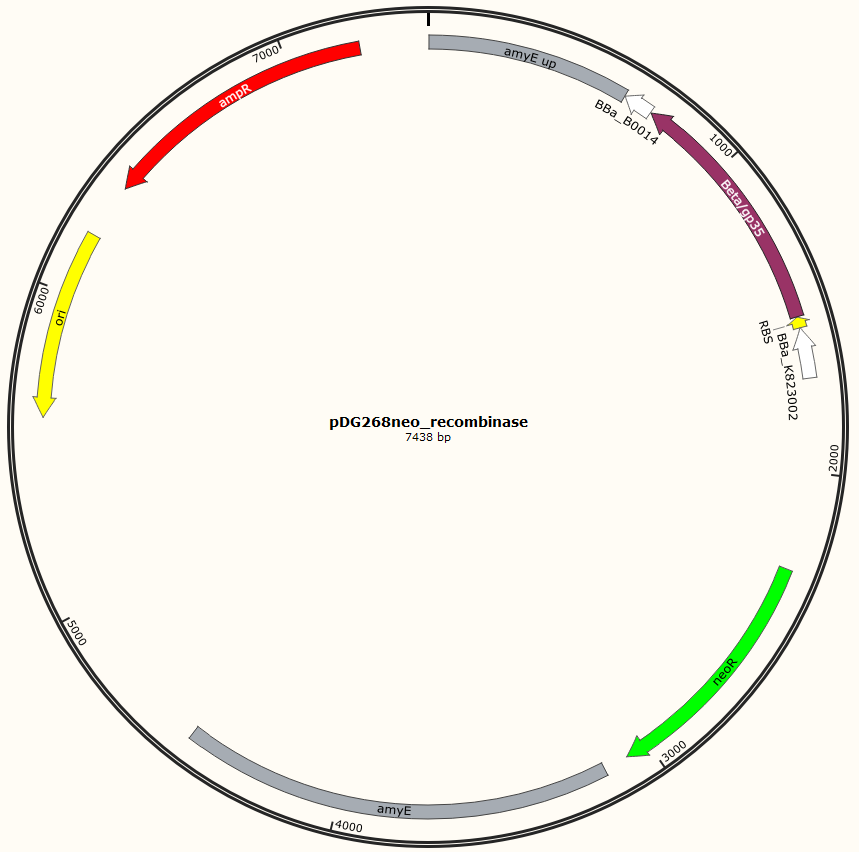
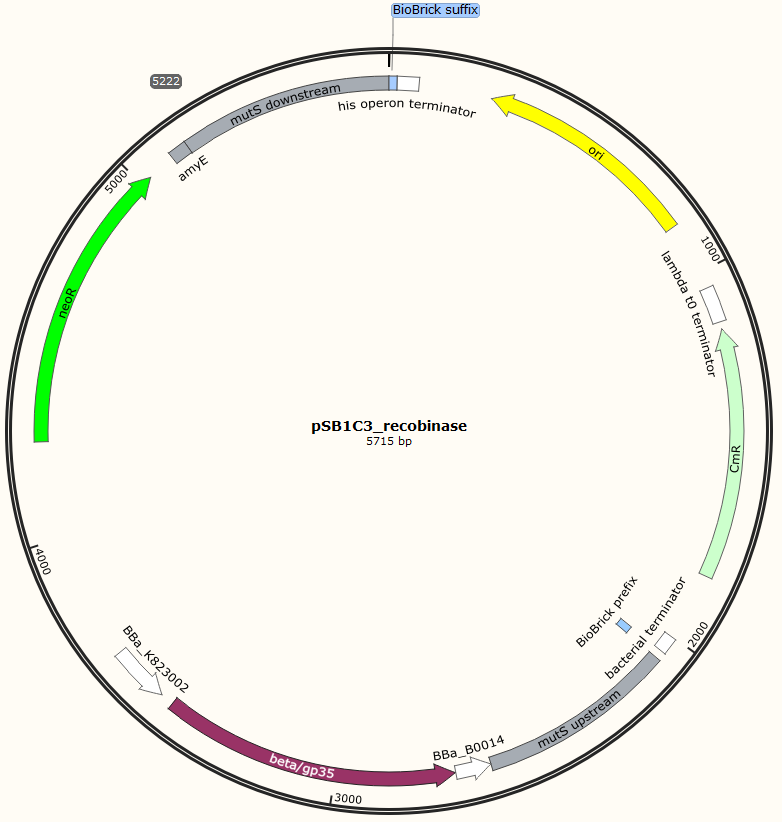
Four different plasmids was assembled to make the four MAGE ready strains:
pDG268neo_Beta-neoR
pDG268neo_GP35-neoR
pSB1C3_Beta-neoR
pSB1C3_GP35-neoR
Missing: figure tekst: Shows the general concepts of the two plasmids pDG268neo_recombinase and pSB1C3_recombinase. Both exists in two versions, one with each of the recombinase proteins CDSs (Beta and GP35). Those also have different RBSs since they are optimized for the CDS. Upstream of neoR is a promoter and RBS and downstream of neoR is a terminator, but sequences and positions of these features are not known.
For pDG268neo_recobinase a DNA sequence containing following features
Missing: make a table on following
● Promoter: PliaG from BBa_K823002 was used.
● RBSs were optimized for the specific CDS using the salis lab RBS calculator (Missing https://www.denovodna.com/software/)
● CDS for recombination protein beta or GP35
● Terminator: we use rho-independent Part:BBa_B0014
Sequence was ordered from IDT as two gblocks (for each recombinase) with overlapping regions, thus they can be assembled with Gibsom assembly.
pDG268neo was linearized in a PCR with primers, so that the native lacZ was omitted. This linearized plasmid was purified and used in a Gibsom assembly reaction with two cognate gblocks. This is shown on the figure missing.
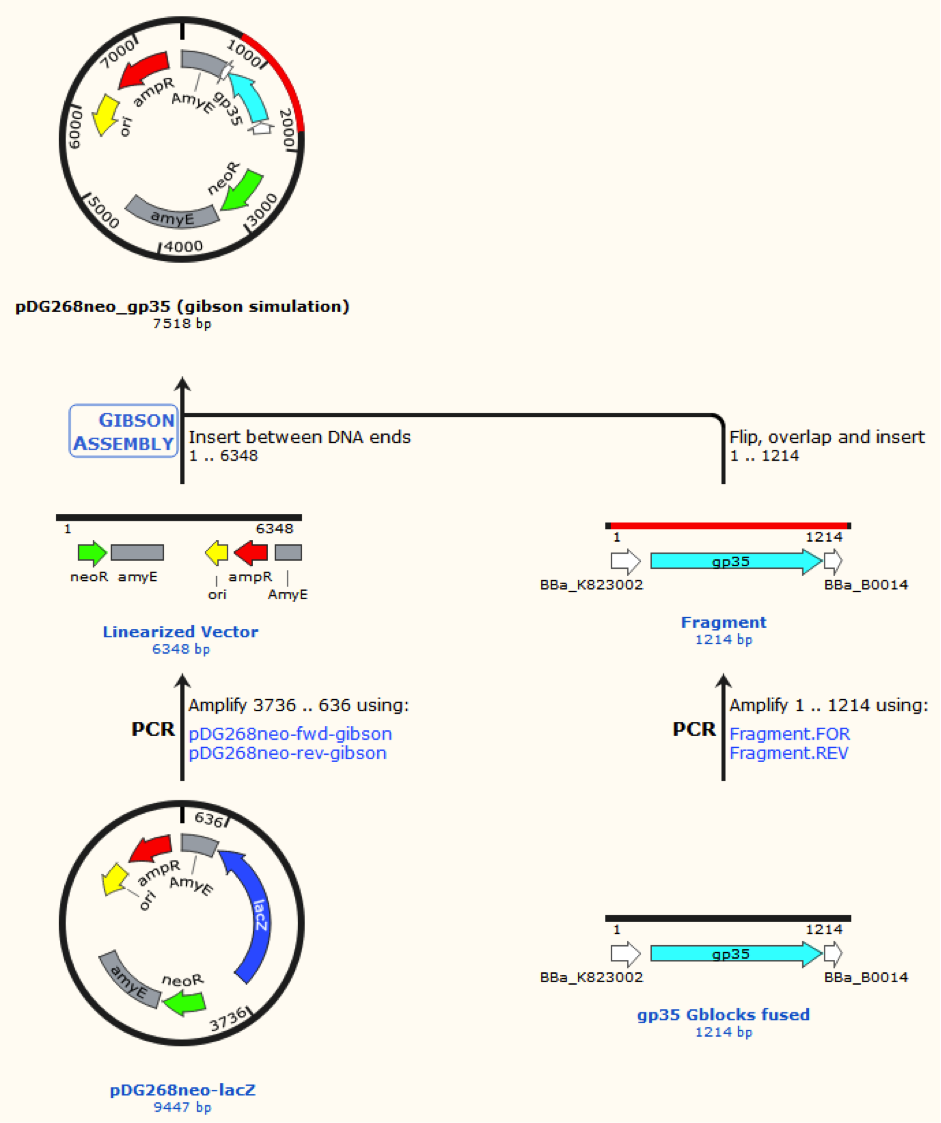
Missing figure text: Gibsom assembly of the pDG268neo_GP35, the gblocks was fused to the “gp35 Gblocks fused” in the same reaction. The Gibsom assembly of pDG268neo_beta was similar.
This resulted in two different plasmids pDG268neo_beta and pDG268neo_gp35.
The two mutS KO plasmids was made of the following DNA fragments:
1. About 500 bp up- and downstream from mutS was amplified, using primers with tails.
2. Recombinase and neoR expression cassettes was amplified from the pDG268neo_recombinase
3. Linearized pSB1C3 was used as template.
These fragments was assembled into two different plasmids pSB1C3_mutS::beta-neoR and pSB1C3_mutS::gp35-neoR using Gibsom assembly.
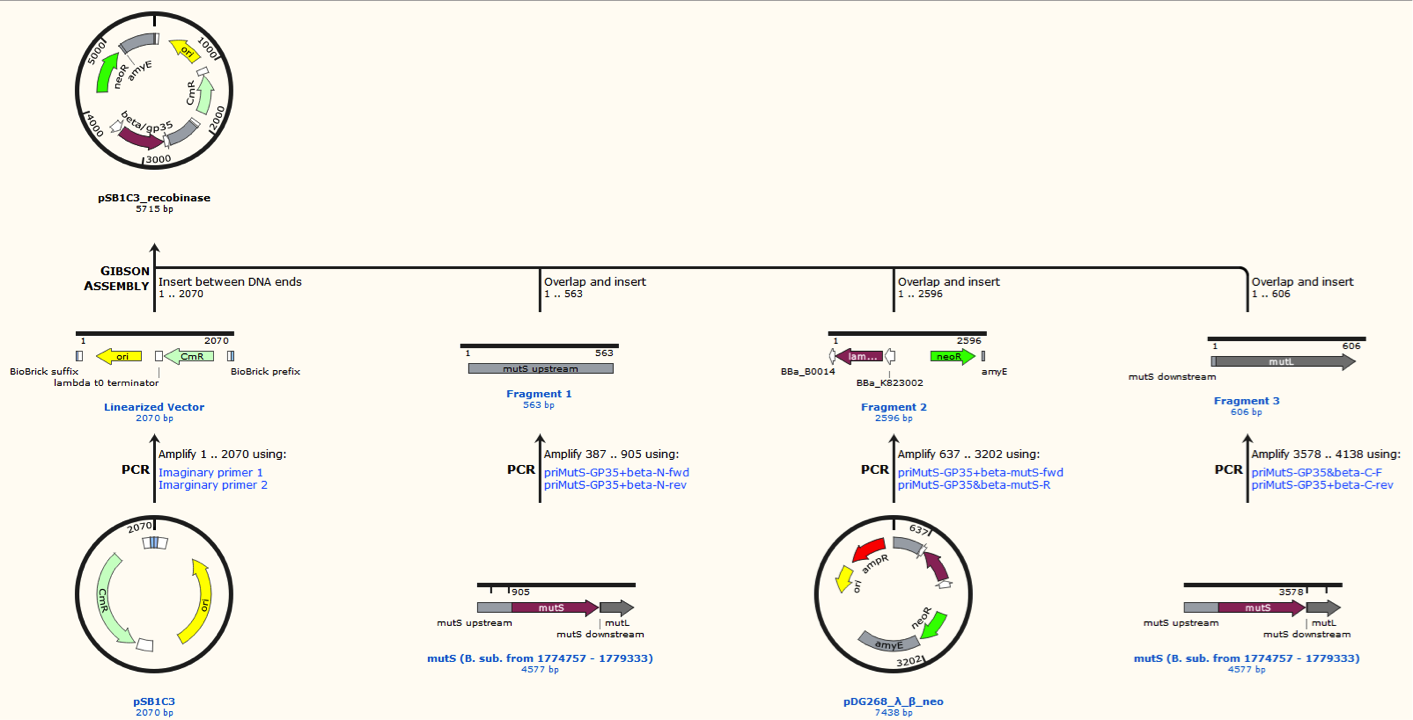
Missing figure text: Gibsom assembly of the pSB1C3_beta. The feature called “mutL” corresponding mutS downstream. The Gibsom assembly of pSB1C3_GP35 was similar.
All plasmids were verified by restriction enzyme digestion.
We prepared naturel competent B. subtilis 168 to be transformed. The plasmids were linearized with restriction enzymes. Then transformed into naturally competent B. subtilis 168, transformants were selected on 5ɣ neo. Transformants were verified with colony PCRs.
Department of Systems Biology
Søltofts Plads 221
2800 Kgs. Lyngby
Denmark
P: +45 45 25 25 25
M: dtu-igem-2015@googlegroups.com