Difference between revisions of "Team:Toronto/Parts"
| (19 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | <!-- ####################################################### --> | ||
| + | <!-- # This html was produced by the igemwiki generator # --> | ||
| + | <!-- # https://github.com/igemuoftATG/generator-igemwiki # --> | ||
| + | <!-- ####################################################### --> | ||
| + | |||
| + | <!-- repo for this wiki: https://github.com/igemuoftATG/wiki2015 --> | ||
| + | |||
| + | </html> | ||
{{Toronto/head}} | {{Toronto/head}} | ||
<html> | <html> | ||
| − | < | + | <div id="navigation"> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<ul> | <ul> | ||
| − | <li> | + | <li><a href="https://2015.igem.org/Team:Toronto"><span>home</span><i class="fa fa-home"></i></a></li> |
| − | <li> | + | </li> |
| − | <li> | + | <li><a href="https://2015.igem.org/Team:Toronto/Team"><span>team</span><i class="fa fa-users"></i></a></li> |
| − | <li> | + | </li> |
| − | <li> | + | <li><a href="#"><span>project</span><i class="fa fa-info-circle"></i></a> |
| − | <li> | + | <ul> |
| − | <li>Design | + | <li><a href="https://2015.igem.org/Team:Toronto/Description"><span>description</span></a></li> |
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Experiments"><span>experiments</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Results"><span>results</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Design"><span>design</span></a></li> | ||
| + | </li> | ||
</ul> | </ul> | ||
| − | < | + | <li class="active"><a href="https://2015.igem.org/Team:Toronto/Parts"><span>parts</span><i class="fa fa-gear"></i></a></li> |
| − | + | </li> | |
| − | + | <li><a href="https://2015.igem.org/Team:Toronto/Notebook"><span>notebook</span><i class="fa fa-book"></i></a></li> | |
| − | < | + | </li> |
| − | < | + | <li><a href="https://2015.igem.org/Team:Toronto/Attributions"><span>attributions</span><i class="fa fa-life-saver"></i></a></li> |
| − | + | </li> | |
| − | + | <li><a href="https://2015.igem.org/Team:Toronto/Collaborations"><span>collaborations</span><i class="fa fa-users"></i></a></li> | |
| − | < | + | </li> |
| + | <li><a href="#"><span>practices</span><i class="fa fa-tree"></i></a> | ||
<ul> | <ul> | ||
| − | <li><a href="https:// | + | <li><a href="https://2015.igem.org/Team:Toronto/Practices"><span>human practices</span></a></li> |
| − | <li><a href="https:// | + | </li> |
| − | <li><a href="https:// | + | <li><a href="https://2015.igem.org/Team:Toronto/Alberta-Oil-Stories"><span>alberta oil stories</span></a></li> |
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Community-Outreach"><span>community outreach</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Technology-Applications"><span>technology applications</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/TechnoMoral-Scenarios"><span>technoMoral scenarios</span></a></li> | ||
| + | </li> | ||
</ul> | </ul> | ||
| − | < | + | <li><a href="https://2015.igem.org/Team:Toronto/Safety"><span>safety</span><i class="fa fa-fire"></i></a></li> |
| − | < | + | </li> |
| − | < | + | <li><a href="#"><span>software</span><i class="fa fa-code"></i></a> |
| − | <html> | + | <ul> |
| + | <li><a href="https://2015.igem.org/Team:Toronto/Software"><span>software</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Demo"><span>demo</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Generator"><span>wiki generator</span></a></li> | ||
| + | </li> | ||
| + | </ul> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Entrepreneurship"><span>entrepreneurship</span><i class="fa fa-institution"></i></a></li> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="content" id="content-main"><div class="row"><div class="col col-lg-8 col-md-12"><div class="content-main"><h1 id="toluene-degradation-and-our-project">Toluene Degradation and Our Project</h1> | ||
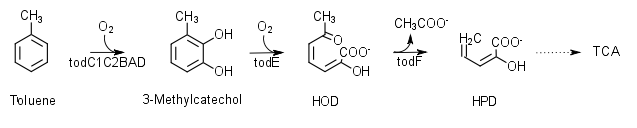
| + | <p>Pseudomonas putida F1 species degrades toluene with the help of the tod operon. Toluene dioxygenase and toluene cis-dihydrodiol dehydroxygenase transcribed from todC1C2BA and todD genes, respectively, facilitate the breakdown of toluene to 3-methylcatechol. Gerben J. Zylstra et al. successfully constructed the plasmid pTDG602 containing todC1C2BAD genes. To achieve complete degradation of toluene down to carbon dioxide and water, we focused on designing a plasmid containing todE and todF genes. After cotransformation of DH10B cells with pTDG602 and our plasmid, E.Coli should theoretically contain all the genes required to achieve complete degradation.</p> | ||
| + | <p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_Pathway.png]] <html> | ||
| + | </p> | ||
| + | <h2 id="submitted-part-tode-biobrick-bba_k1760001-">Submitted Part: TodE BioBrick (BBa_K1760001)</h2> | ||
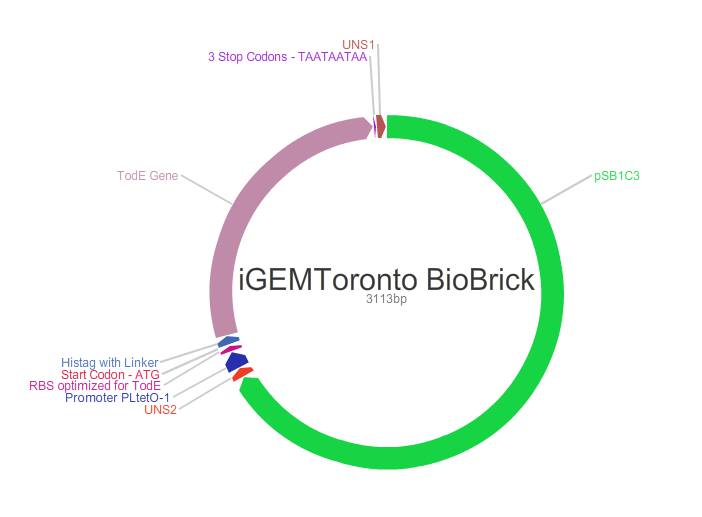
| + | <p>Final submitted biobrick is a coding sequence that contains a TetO promoter, RBS optimized by using Salis RBS caluclator and codon optimized todE gene for E.Coli. It plays a major role in degradation of toluene. Specifically, todE degrades 3-methylcatechol, a highly toxic intermediate into HOD. In pseudomonas putida species, HOD is aerobically metabolized through TCA cycle. Theoretically, a E.Coli MG1655 type bacteria containing todC1C2BADEF genes can degrade toluene through TCA cycle by using its own native enzymes.</p> | ||
| + | <p>This part is ordered in gene blocks and ligated by using Gibson Assembly by using . The design have been performed by Anthony Zhao, Seray Cicek, Umar Owadally, Katariina Jaenes, Matt D'lorio, ChungMin Lee, Drupad Rajyaguru who are wet lab team members of the iGEM Toronto 2015 team. See our part in the the registry: <a href="http://parts.igem.org/Part:BBa_K1760001">http://parts.igem.org/Part:BBa_K1760001</a></p> | ||
| + | <p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_TodE-plasmid_new.jpg]] <html> | ||
| + | </p> | ||
| + | <h2 id="additional-designs-in-progress">Additional Designs in Progress</h2> | ||
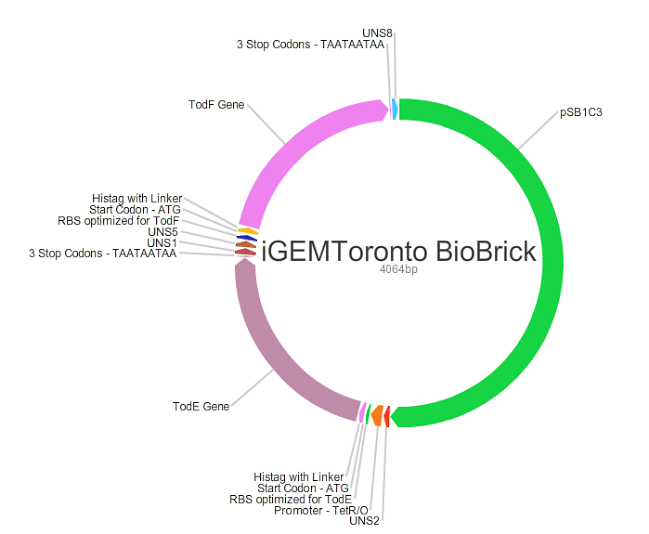
| + | <p>In order to achieve complete degradation of toluene, we also constructed a codon optimized todF sequence. We tried to create a ligated plasmid containing todE, todF, optimized Salis RBS calculator and a tunable promoter to determine optimum transcription for DH10B cells. We used Ligase Cycling Reaction(LCR) in this process. Unique Nucleotide Sequences are added to ligating blocks to facilitate assembly of the plasmid without a secondary structure interference.</p> | ||
| + | <p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_biobrick_plasmid.png]] <html> | ||
| + | </p> | ||
| + | |||
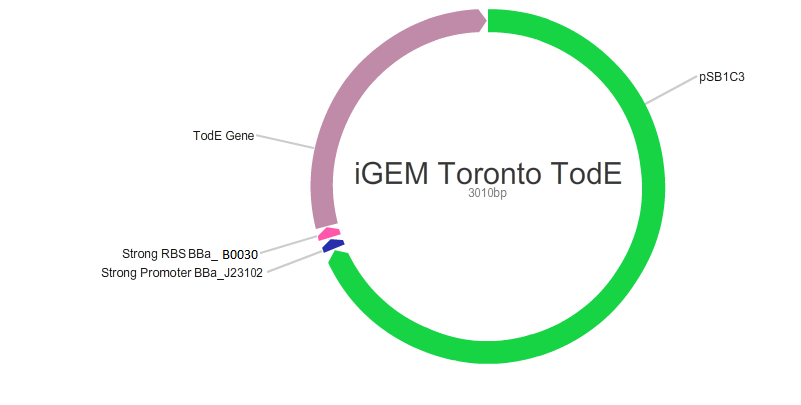
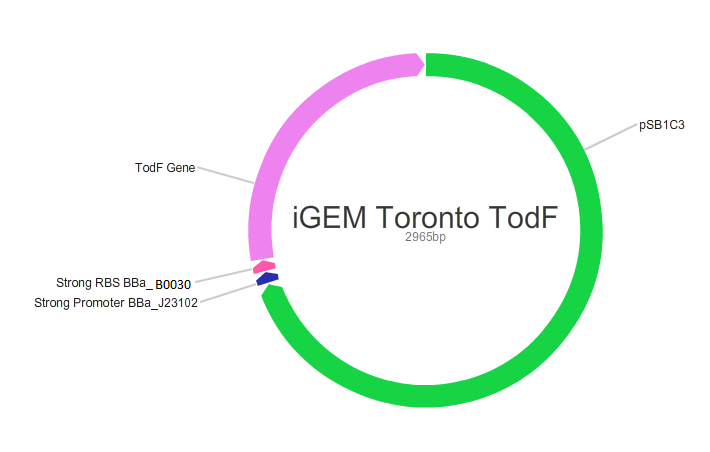
| + | <p>We also used BioBricks from the registry to design plasmids with different efficiencies by using RDP kit as shown below. In this process, we used promoter BBa_J23102 and RBS BBa_B0030.</p> | ||
| + | <p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_TodEplasmid.png]] <html> | ||
| + | </p> | ||
| + | |||
| + | <p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_TodFplasmid.png]] <html> | ||
| + | </p> | ||
| + | |||
| + | <p>Note: Links to iGem registry | ||
| + | BBa_J23102: <a href="http://parts.igem.org/Part:BBa_J23102">http://parts.igem.org/Part:BBa_J23102</a> | ||
| + | BBa_0030: <a href="http://parts.igem.org/Part:BBa_B0030">http://parts.igem.org/Part:BBa_B0030</a></p> | ||
| + | <ul> | ||
| + | <li>Sequence for BBa_K1760001 :</li> | ||
| + | <li>tactagtagcggccgctgcagtccggcaaaaaagggcaaggtgtcaccaccctgccctttttctttaaaaccgaaaagattacttcgcgttatgcaggct | ||
| + | tcctcgctcactgactcgctgcgctcggtcgttcggctgcggcgagcggtatcagctcactcaaaggcggtaatacggttatccacagaatcaggggata | ||
| + | acgcaggaaagaacatgtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccgcgttgctggcgtttttccacaggctccgcccccctgacga | ||
| + | gcatcacaaaaatcgacgctcaagtcagaggtggcgaaacccgacaggactataaagataccaggcgtttccccctggaagctccctcgtgcgctctcct | ||
| + | gttccgaccctgccgcttaccggatacctgtccgcctttctcccttcgggaagcgtggcgctttctcatagctcacgctgtaggtatctcagttcggtgt | ||
| + | aggtcgttcgctccaagctgggctgtgtgcacgaaccccccgttcagcccgaccgctgcgccttatccggtaactatcgtcttgagtccaacccggtaag | ||
| + | acacgacttatcgccactggcagcagccactggtaacaggattagcagagcgaggtatgtaggcggtgctacagagttcttgaagtggtggcctaactac | ||
| + | ggctacactagaagaacagtatttggtatctgcgctctgctgaagccagttaccttcggaaaaagagttggtagctcttgatccggcaaacaaaccaccg | ||
| + | ctggtagcggtggtttttttgtttgcaagcagcagattacgcgcagaaaaaaaggatctcaagaagatcctttgatcttttctacggggtctgacgctca | ||
| + | gtggaacgaaaactcacgttaagggattttggtcatgagattatcaaaaaggatcttcacctagatccttttaaattaaaaatgaagttttaaatcaatc | ||
| + | taaagtatatatgagtaaacttggtctgacagctcgaggcttggattctcaccaataaaaaacgcccggcggcaaccgagcgttctgaacaaatccagat | ||
| + | ggagttctgaggtcattactggatctatcaacaggagtccaagcgagctcgatatcaaattacgccccgccctgccactcatcgcagtactgttgtaatt | ||
| + | cattaagcattctgccgacatggaagccatcacaaacggcatgatgaacctgaatcgccagcggcatcagcaccttgtcgccttgcgtataatatttgcc | ||
| + | catggtgaaaacgggggcgaagaagttgtccatattggccacgtttaaatcaaaactggtgaaactcacccagggattggctgagacgaaaaacatattc | ||
| + | tcaataaaccctttagggaaataggccaggttttcaccgtaacacgccacatcttgcgaatatatgtgtagaaactgccggaaatcgtcgtggtattcac | ||
| + | tccagagcgatgaaaacgtttcagtttgctcatggaaaacggtgtaacaagggtgaacactatcccatatcaccagctcaccgtctttcattgccatacg | ||
| + | aaattccggatgagcattcatcaggcgggcaagaatgtgaataaaggccggataaaacttgtgcttatttttctttacggtctttaaaaaggccgtaata | ||
| + | tccagctgaacggtctggttataggtacattgagcaactgactgaaatgcctcaaaatgttctttacgatgccattgggatatatcaacggtggtatatc | ||
| + | cagtgatttttttctccattttagcttccttagctcctgaaaatctcgataactcaaaaaatacgcccggtagtgatcttatttcattatggtgaaagtt | ||
| + | ggaacctcttacgtgcccgatcaactcgagtgccacctgacgtctaagaaaccattattatcatgacattaacctataaaaataggcgtatcacgaggca | ||
| + | gaatttcagataaaaaaaatccttagctttcgctaaggatgatttctggaattcgcgggctgggagttcgtagacggaaacaaacgcatccctatcagtg | ||
| + | atagagattgacatccctatcagtgatagagatactgagcacacggataaggaggtaagttatgcatcaccaccatcaccatggcagtggcagcattcag | ||
| + | aggctaggctatctaggattcgaagtcgccgatgtgcgctcctggaggaccttcgcgaccacccgcctaggcatgatggaagccagcgcaagcgaaaccg | ||
| + | aagcgacctttcgcattgatagccgcgcttggcgcctcagcgtcagccgcggcccggctgatgattatctgttcgcgggttttgaagtggatagcgaaca | ||
| + | aggccttcaggaagtgaaagaaagcctccaggcgcatggcgtcaccgtgaaagtggaaggcggcgaactgattgctaaacgcggcgtgctgggtctgatc | ||
| + | agctgcaccgatccgtttggcaaccgcgtggaaatttattacggtgcgaccgaactgtttgaacgcccgttcgcgagcccgacaggcgtgagtggttttc | ||
| + | agaccggcgatcagggactgggccattatgtgctaagcgtggcagatgtggatgcggcgctggcgttttataccaaagcgctgggctttcagctggcgga | ||
| + | tgtgattgattggaccattggcgatggcctgagcgtgaccctgtattttctgtattgcaacgggcgccatcatagcttcgcgtttgcgaaactgccgggc | ||
| + | agcaaacgcctgcatcattttatgctccaggcgaacggcatggatgatgtgggcctggcgtatgataagtttgatgcggaacgcgcggtggtgatgagcc | ||
| + | tgggccgccataccaacgatcatatgattagcttttatggcgcgaccccgagcggctttgcggtggaatatggctggggcgcgcgcgaagtgacccgcca | ||
| + | ttggagcgtggtgcgctatgatcgcattagcatttggggccataaatttcaggcgccggcgtaataataacattactcgcatccattctcaggctgtctc | ||
| + | g</li> | ||
| + | </ul> | ||
| + | </div></div><div id="tableofcontents" class="tableofcontents affix sidebar col-lg-4 hidden-xs hidden-sm hidden-md visible-lg-3"><ul class="nav"> | ||
| + | <li><a href="#submitted-part--tode-biobrick--bba-k1760001-">Submitted Part: TodE BioBrick (BBa_K1760001)</a></li> | ||
| + | <li><a href="#additional-designs-in-progress">Additional Designs in Progress</a></li> | ||
| + | </ul> | ||
| + | </div></div></div> | ||
</html> | </html> | ||
{{Toronto/footer}} | {{Toronto/footer}} | ||
Latest revision as of 04:30, 20 November 2015
Toluene Degradation and Our Project
Pseudomonas putida F1 species degrades toluene with the help of the tod operon. Toluene dioxygenase and toluene cis-dihydrodiol dehydroxygenase transcribed from todC1C2BA and todD genes, respectively, facilitate the breakdown of toluene to 3-methylcatechol. Gerben J. Zylstra et al. successfully constructed the plasmid pTDG602 containing todC1C2BAD genes. To achieve complete degradation of toluene down to carbon dioxide and water, we focused on designing a plasmid containing todE and todF genes. After cotransformation of DH10B cells with pTDG602 and our plasmid, E.Coli should theoretically contain all the genes required to achieve complete degradation.
Submitted Part: TodE BioBrick (BBa_K1760001)
Final submitted biobrick is a coding sequence that contains a TetO promoter, RBS optimized by using Salis RBS caluclator and codon optimized todE gene for E.Coli. It plays a major role in degradation of toluene. Specifically, todE degrades 3-methylcatechol, a highly toxic intermediate into HOD. In pseudomonas putida species, HOD is aerobically metabolized through TCA cycle. Theoretically, a E.Coli MG1655 type bacteria containing todC1C2BADEF genes can degrade toluene through TCA cycle by using its own native enzymes.
This part is ordered in gene blocks and ligated by using Gibson Assembly by using . The design have been performed by Anthony Zhao, Seray Cicek, Umar Owadally, Katariina Jaenes, Matt D'lorio, ChungMin Lee, Drupad Rajyaguru who are wet lab team members of the iGEM Toronto 2015 team. See our part in the the registry: http://parts.igem.org/Part:BBa_K1760001
Additional Designs in Progress
In order to achieve complete degradation of toluene, we also constructed a codon optimized todF sequence. We tried to create a ligated plasmid containing todE, todF, optimized Salis RBS calculator and a tunable promoter to determine optimum transcription for DH10B cells. We used Ligase Cycling Reaction(LCR) in this process. Unique Nucleotide Sequences are added to ligating blocks to facilitate assembly of the plasmid without a secondary structure interference.
We also used BioBricks from the registry to design plasmids with different efficiencies by using RDP kit as shown below. In this process, we used promoter BBa_J23102 and RBS BBa_B0030.
Note: Links to iGem registry BBa_J23102: http://parts.igem.org/Part:BBa_J23102 BBa_0030: http://parts.igem.org/Part:BBa_B0030
- Sequence for BBa_K1760001 :
- tactagtagcggccgctgcagtccggcaaaaaagggcaaggtgtcaccaccctgccctttttctttaaaaccgaaaagattacttcgcgttatgcaggct tcctcgctcactgactcgctgcgctcggtcgttcggctgcggcgagcggtatcagctcactcaaaggcggtaatacggttatccacagaatcaggggata acgcaggaaagaacatgtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccgcgttgctggcgtttttccacaggctccgcccccctgacga gcatcacaaaaatcgacgctcaagtcagaggtggcgaaacccgacaggactataaagataccaggcgtttccccctggaagctccctcgtgcgctctcct gttccgaccctgccgcttaccggatacctgtccgcctttctcccttcgggaagcgtggcgctttctcatagctcacgctgtaggtatctcagttcggtgt aggtcgttcgctccaagctgggctgtgtgcacgaaccccccgttcagcccgaccgctgcgccttatccggtaactatcgtcttgagtccaacccggtaag acacgacttatcgccactggcagcagccactggtaacaggattagcagagcgaggtatgtaggcggtgctacagagttcttgaagtggtggcctaactac ggctacactagaagaacagtatttggtatctgcgctctgctgaagccagttaccttcggaaaaagagttggtagctcttgatccggcaaacaaaccaccg ctggtagcggtggtttttttgtttgcaagcagcagattacgcgcagaaaaaaaggatctcaagaagatcctttgatcttttctacggggtctgacgctca gtggaacgaaaactcacgttaagggattttggtcatgagattatcaaaaaggatcttcacctagatccttttaaattaaaaatgaagttttaaatcaatc taaagtatatatgagtaaacttggtctgacagctcgaggcttggattctcaccaataaaaaacgcccggcggcaaccgagcgttctgaacaaatccagat ggagttctgaggtcattactggatctatcaacaggagtccaagcgagctcgatatcaaattacgccccgccctgccactcatcgcagtactgttgtaatt cattaagcattctgccgacatggaagccatcacaaacggcatgatgaacctgaatcgccagcggcatcagcaccttgtcgccttgcgtataatatttgcc catggtgaaaacgggggcgaagaagttgtccatattggccacgtttaaatcaaaactggtgaaactcacccagggattggctgagacgaaaaacatattc tcaataaaccctttagggaaataggccaggttttcaccgtaacacgccacatcttgcgaatatatgtgtagaaactgccggaaatcgtcgtggtattcac tccagagcgatgaaaacgtttcagtttgctcatggaaaacggtgtaacaagggtgaacactatcccatatcaccagctcaccgtctttcattgccatacg aaattccggatgagcattcatcaggcgggcaagaatgtgaataaaggccggataaaacttgtgcttatttttctttacggtctttaaaaaggccgtaata tccagctgaacggtctggttataggtacattgagcaactgactgaaatgcctcaaaatgttctttacgatgccattgggatatatcaacggtggtatatc cagtgatttttttctccattttagcttccttagctcctgaaaatctcgataactcaaaaaatacgcccggtagtgatcttatttcattatggtgaaagtt ggaacctcttacgtgcccgatcaactcgagtgccacctgacgtctaagaaaccattattatcatgacattaacctataaaaataggcgtatcacgaggca gaatttcagataaaaaaaatccttagctttcgctaaggatgatttctggaattcgcgggctgggagttcgtagacggaaacaaacgcatccctatcagtg atagagattgacatccctatcagtgatagagatactgagcacacggataaggaggtaagttatgcatcaccaccatcaccatggcagtggcagcattcag aggctaggctatctaggattcgaagtcgccgatgtgcgctcctggaggaccttcgcgaccacccgcctaggcatgatggaagccagcgcaagcgaaaccg aagcgacctttcgcattgatagccgcgcttggcgcctcagcgtcagccgcggcccggctgatgattatctgttcgcgggttttgaagtggatagcgaaca aggccttcaggaagtgaaagaaagcctccaggcgcatggcgtcaccgtgaaagtggaaggcggcgaactgattgctaaacgcggcgtgctgggtctgatc agctgcaccgatccgtttggcaaccgcgtggaaatttattacggtgcgaccgaactgtttgaacgcccgttcgcgagcccgacaggcgtgagtggttttc agaccggcgatcagggactgggccattatgtgctaagcgtggcagatgtggatgcggcgctggcgttttataccaaagcgctgggctttcagctggcgga tgtgattgattggaccattggcgatggcctgagcgtgaccctgtattttctgtattgcaacgggcgccatcatagcttcgcgtttgcgaaactgccgggc agcaaacgcctgcatcattttatgctccaggcgaacggcatggatgatgtgggcctggcgtatgataagtttgatgcggaacgcgcggtggtgatgagcc tgggccgccataccaacgatcatatgattagcttttatggcgcgaccccgagcggctttgcggtggaatatggctggggcgcgcgcgaagtgacccgcca ttggagcgtggtgcgctatgatcgcattagcatttggggccataaatttcaggcgccggcgtaataataacattactcgcatccattctcaggctgtctc g