Difference between revisions of "Team:UESTC Software/Collaborations.html"
(Created page with "<!DOCTYPE html> <html class="no-js"> <head> <meta charset="UTF-8"> <meta name="viewport" content="width=device-width, initial-scale=1"> <title>Meet Up - Human Practice</title>...") |
|||
| Line 121: | Line 121: | ||
<div class="footer_define"> | <div class="footer_define"> | ||
<div class="footer_logo"> | <div class="footer_logo"> | ||
| − | <img src=" | + | <img src="/wiki/images/0/07/Uestc_software_self_logo.png"> |
<p>@2015 Designed & Developed by <span class="green_em">UESTC_SOFTWARE</span> iGEM</p> | <p>@2015 Designed & Developed by <span class="green_em">UESTC_SOFTWARE</span> iGEM</p> | ||
</div> | </div> | ||
Revision as of 17:27, 27 August 2015
<!DOCTYPE html>
Meet-up
In August, we cooperated with SCU_China iGEM team to study how to find out SD sequence in DNA sequence and analyze the strength of the SD sequence that affects translation issues. Then we developed an online software based on browser—SD finder (Figure 1), which is used in helping user predict the ORF and SD sequence of DNA sequence.
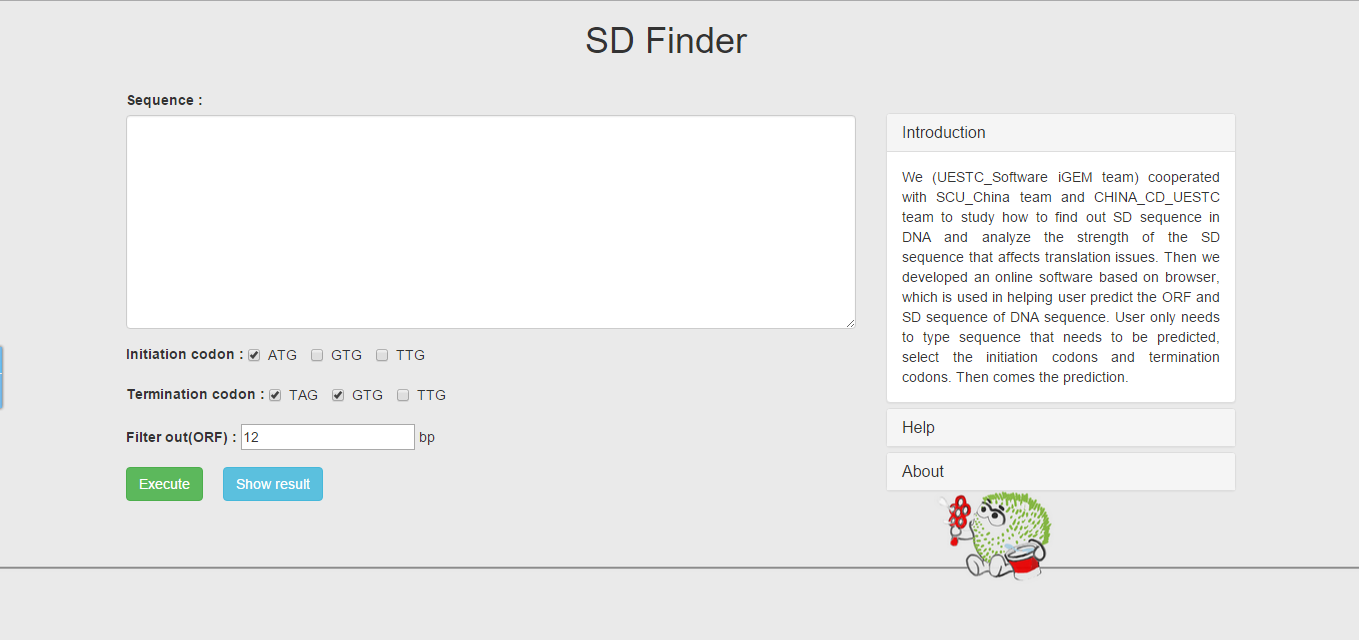
Figure 1 online software based on browser—SD finder
Though the SD finder doesn`t require the length of the sequence for predicting, we suggest that user use short sequence (hundreds of bp ~ a few KB). User only needs to type sequence that needs to be predicted, select the start codons and stop codons. Then comes the prediction.
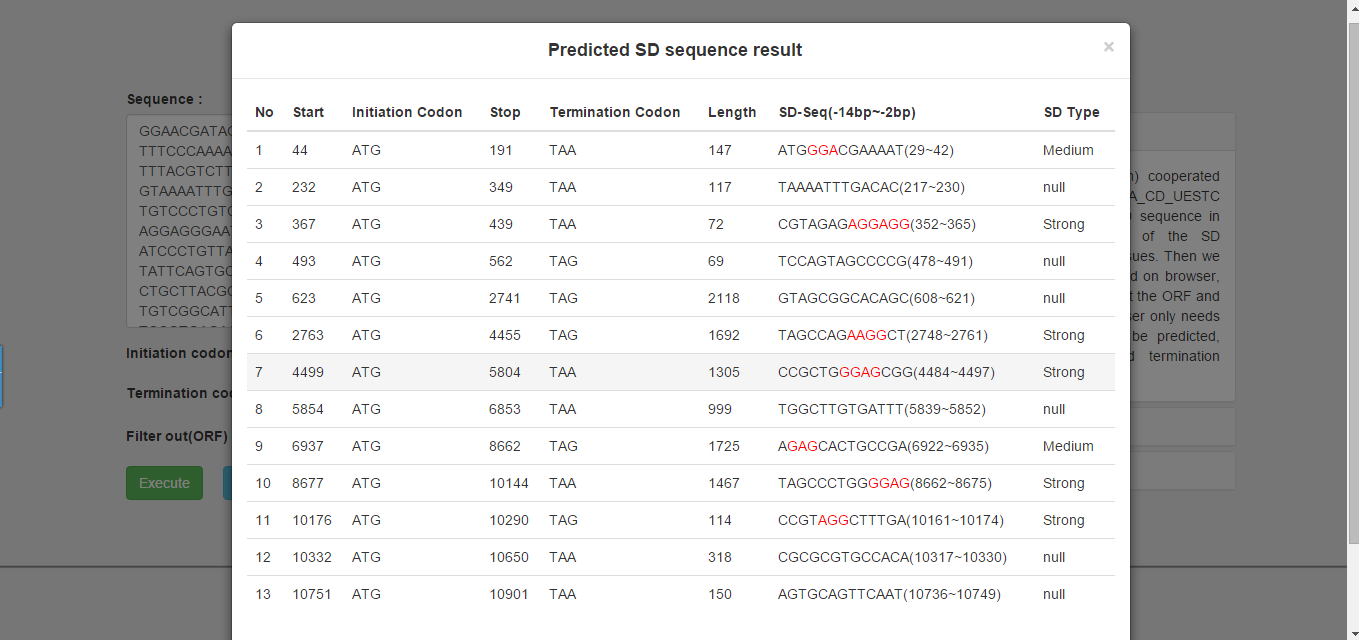
We provide the related information about ORF and SD sequence in the result page (Figure 2). The column of SD sequence lists a 13bp sequence from -2 to -14 in ORF upstream. We also tag the position of SD sequence signals with red. And the last column lists the strength of the type of the SD sequence.