Difference between revisions of "Team:UESTC Software/Design.html"
| Line 65: | Line 65: | ||
<div class="safetyDiv"> | <div class="safetyDiv"> | ||
<div class="QandA"> | <div class="QandA"> | ||
| − | <h3 class=" | + | <h3 class="design_h3">Project Base</h3> |
<p>Because of its widespread value of application and potential, the study of essential genes has been concerned by more and more synthetic biologists and researchers. As more and more bacterial genes have been identified as essential genes, some databases like DEG (Database of Essential Genes), OGEE (Online Gene Essentiality database) have appeared. DEG, which is the first essential genes database, only collected essential genes that are tested by whole genome experiments [5]. OGEE not only has essential and nonessential genes, but also has gene characteristic, expressive information and so on [6]. Besides, another database, CEG (Cluster of Essential Genes database), which is set up basing on previous databases, is different from DEG and OGEE. It saves essential genes as homologous cluster. CEG has more information that comes from other databases and is more convenient for studying antibiotics targeting medicine, evolution and etc. | <p>Because of its widespread value of application and potential, the study of essential genes has been concerned by more and more synthetic biologists and researchers. As more and more bacterial genes have been identified as essential genes, some databases like DEG (Database of Essential Genes), OGEE (Online Gene Essentiality database) have appeared. DEG, which is the first essential genes database, only collected essential genes that are tested by whole genome experiments [5]. OGEE not only has essential and nonessential genes, but also has gene characteristic, expressive information and so on [6]. Besides, another database, CEG (Cluster of Essential Genes database), which is set up basing on previous databases, is different from DEG and OGEE. It saves essential genes as homologous cluster. CEG has more information that comes from other databases and is more convenient for studying antibiotics targeting medicine, evolution and etc. | ||
For our project, MCCAP (Minimal Cell Construct and Analysis Panel), which is developed basing on CEG to find “core” essential genes, helps structure minimal gene set and analyse metabolism pathway.</p> | For our project, MCCAP (Minimal Cell Construct and Analysis Panel), which is developed basing on CEG to find “core” essential genes, helps structure minimal gene set and analyse metabolism pathway.</p> | ||
</div> | </div> | ||
<div class="QandA"> | <div class="QandA"> | ||
| − | <h3 class=" | + | <h3 class="design_h3">Theories of MCCAP</h3> |
<p>The major data of MCCAP is derived from CEG. The organizational relationships of the data are shown in the following figure.</p> | <p>The major data of MCCAP is derived from CEG. The organizational relationships of the data are shown in the following figure.</p> | ||
<img class="design_img" src="/wiki/images/8/84/Uestc_software_design.png" /> | <img class="design_img" src="/wiki/images/8/84/Uestc_software_design.png" /> | ||
| Line 76: | Line 76: | ||
</div> | </div> | ||
<div class="QandA"> | <div class="QandA"> | ||
| − | <h3 class=" | + | <h3 class="design_h3">Work flow of screening the minimal gene set</h3> |
<img class="design_img" src="/wiki/images/3/30/Uestc_software_design_1.png"/> | <img class="design_img" src="/wiki/images/3/30/Uestc_software_design_1.png"/> | ||
</div> | </div> | ||
<div class="QandA"> | <div class="QandA"> | ||
| − | <h3 class=" | + | <h3 class="design_h3">Work flow of genetating the pathway and module</h3> |
<img class="design_img" src="/wiki/images/7/76/Uestc_software_design2.png" /> | <img class="design_img" src="/wiki/images/7/76/Uestc_software_design2.png" /> | ||
<p>MACCP creates metabolism pathway by matching K value and using pathway and module database of KEGG. Because of the data that we use refers to the KEGG Orthology (KO) and Cluster of Orthologous Group (COG), not every gene cluster has K value. And we will abandon the gene cluster which doesn`t have K value because it could not create metabolism pathway.</p> | <p>MACCP creates metabolism pathway by matching K value and using pathway and module database of KEGG. Because of the data that we use refers to the KEGG Orthology (KO) and Cluster of Orthologous Group (COG), not every gene cluster has K value. And we will abandon the gene cluster which doesn`t have K value because it could not create metabolism pathway.</p> | ||
Revision as of 12:42, 6 September 2015
<!doctype html>
Design
Project Base
Because of its widespread value of application and potential, the study of essential genes has been concerned by more and more synthetic biologists and researchers. As more and more bacterial genes have been identified as essential genes, some databases like DEG (Database of Essential Genes), OGEE (Online Gene Essentiality database) have appeared. DEG, which is the first essential genes database, only collected essential genes that are tested by whole genome experiments [5]. OGEE not only has essential and nonessential genes, but also has gene characteristic, expressive information and so on [6]. Besides, another database, CEG (Cluster of Essential Genes database), which is set up basing on previous databases, is different from DEG and OGEE. It saves essential genes as homologous cluster. CEG has more information that comes from other databases and is more convenient for studying antibiotics targeting medicine, evolution and etc. For our project, MCCAP (Minimal Cell Construct and Analysis Panel), which is developed basing on CEG to find “core” essential genes, helps structure minimal gene set and analyse metabolism pathway.
Theories of MCCAP
The major data of MCCAP is derived from CEG. The organizational relationships of the data are shown in the following figure.
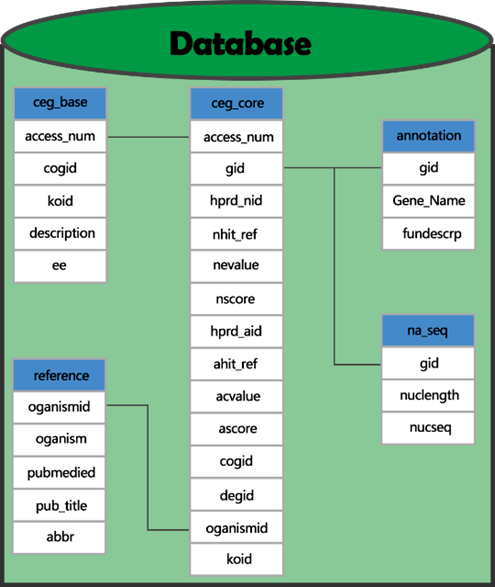
The form of ceg_core and ceg_base represents the core data of essential gene. Each data of form ceg_core is a gene. Data field access_num is a unique ID of gene cluster which has the gene. Gid is a unique ID of genes in Genebank. Koid correspond to the K value of genes in KEGG and cogid correspond to the cog number in COG. And the two numbers may be blank. hprd_nid labeled the highest genetic similarity of genes in Human Protein Reference Database (HPRD). Organismid represents that the gene is from bacterium. Each data in form ceg_base represents a gene cluster. Description is the description of gene cluster function. Ec is the enzyme number of the gene number.
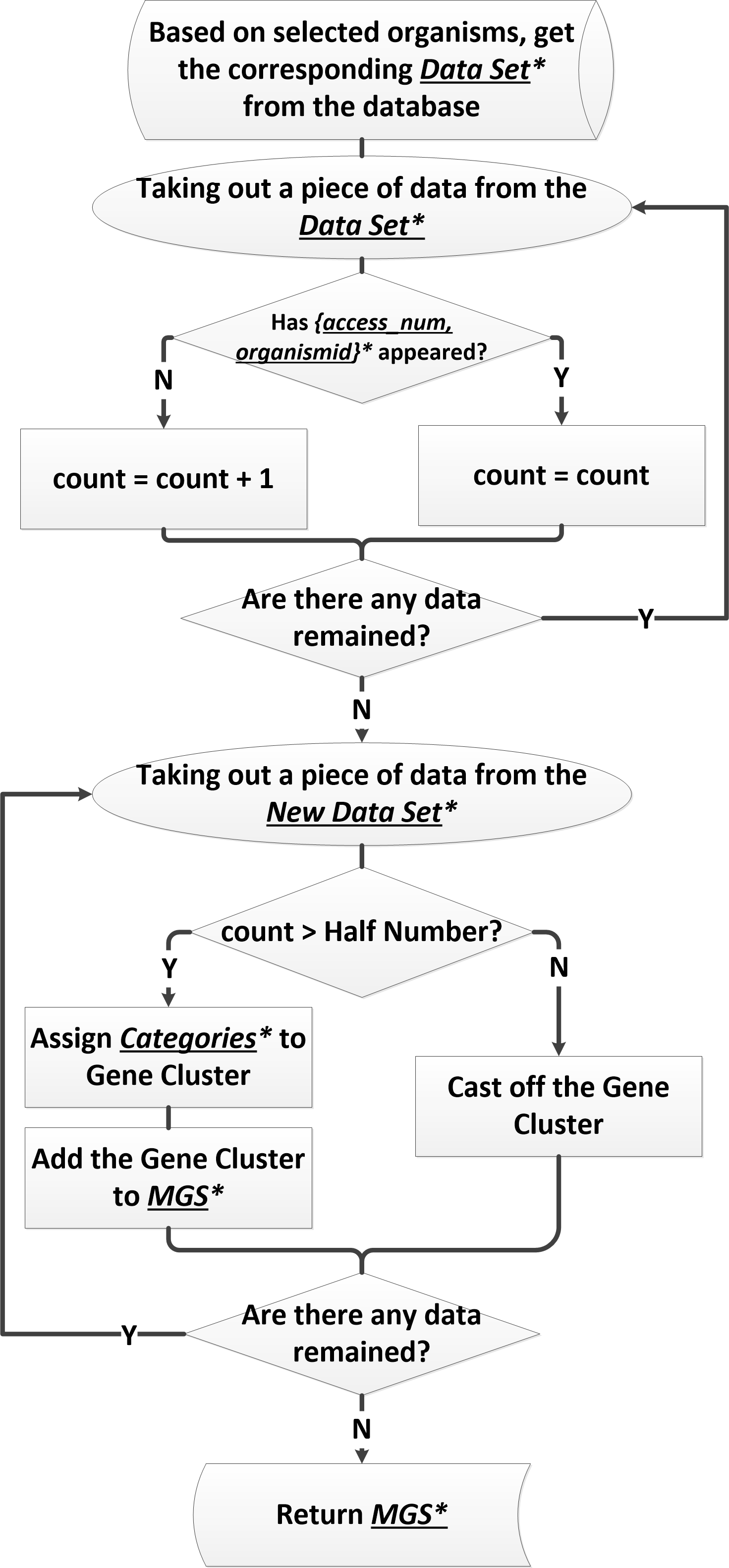
Work flow of screening the minimal gene set
Work flow of genetating the pathway and module
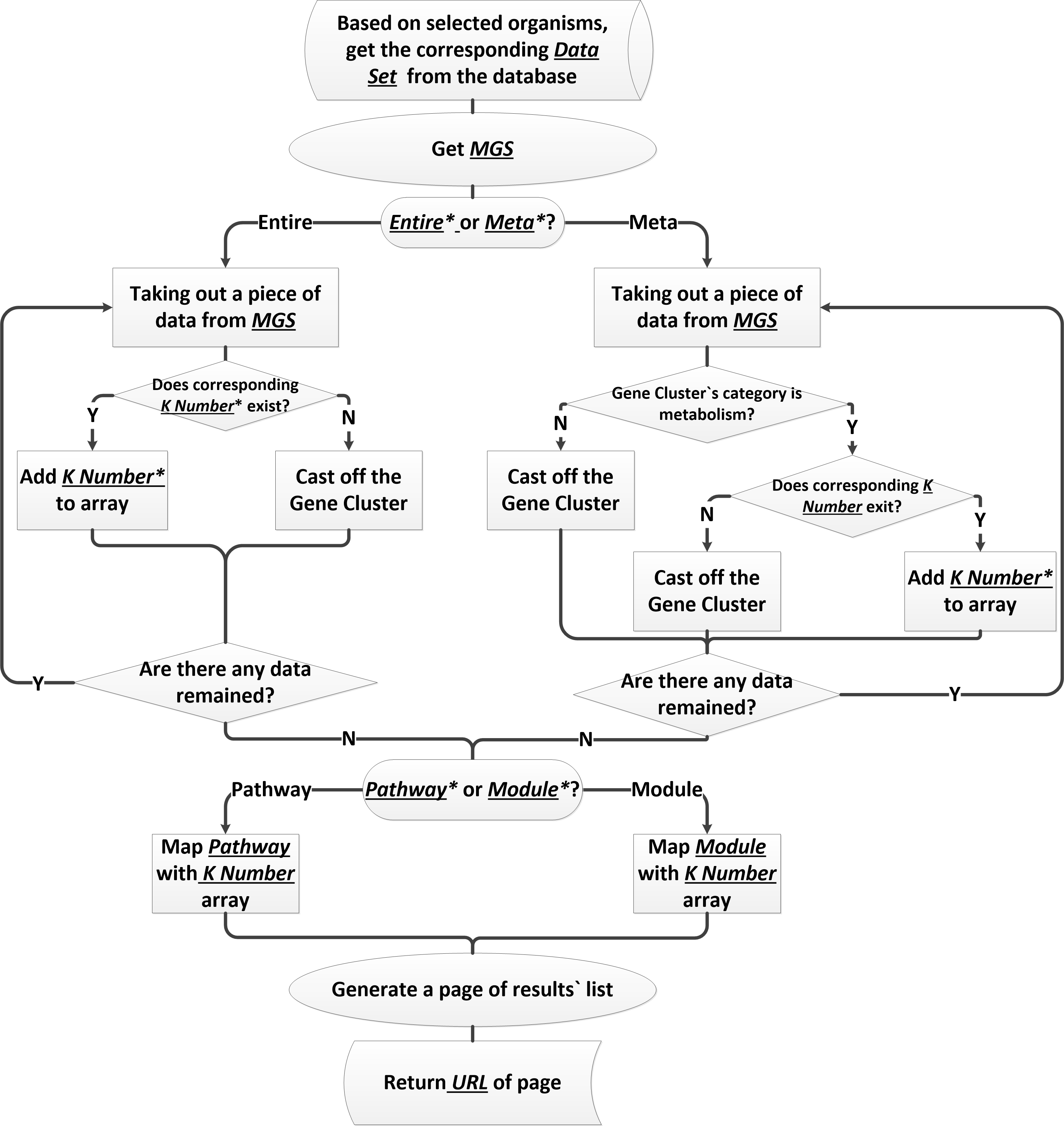
MACCP creates metabolism pathway by matching K value and using pathway and module database of KEGG. Because of the data that we use refers to the KEGG Orthology (KO) and Cluster of Orthologous Group (COG), not every gene cluster has K value. And we will abandon the gene cluster which doesn`t have K value because it could not create metabolism pathway.
