Difference between revisions of "Team:Paris Saclay/Notebook/July/16"
(→Thursday 16th July) |
(→Thursday 16th July) |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Paris_Saclay/notebook_header}} | ||
=Thursday 16th July= | =Thursday 16th July= | ||
| − | + | ==Lab Work== | |
| − | + | ===Plasmid extraction=== | |
| − | ====Plasmid extraction | + | |
''by Johan'' | ''by Johan'' | ||
| − | + | * BBa_S03518 | |
| − | * | + | * BBa_B0015 |
| − | * | + | |
Cultures from the 07/15/2015 | Cultures from the 07/15/2015 | ||
With the Nucleospin Kit from Macherey Nagel | With the Nucleospin Kit from Macherey Nagel | ||
| − | + | ===Digestion=== | |
''by Coralie'' | ''by Coralie'' | ||
| + | |||
| + | * BBa_S03518 | ||
| + | * BBa_B0015 | ||
| + | * BBa_I13602 | ||
In each tube: | In each tube: | ||
| Line 19: | Line 22: | ||
* Buffer FastDigest (10x): 2µL | * Buffer FastDigest (10x): 2µL | ||
* H2O: 6µL | * H2O: 6µL | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Enzymes choice: | Enzymes choice: | ||
| − | * | + | * BBa_S03518 #1: XbaI + PstI |
| − | * | + | * BBa_S03518 #2: SpeI + EcoRI |
| − | * | + | * BBa_B0015 #1: PstI + SpeI |
| − | * | + | * BBa_B0015 #2: XbaI + EcoRI |
| − | * | + | * BBa_I13602 (x2): XbaI + Pst I |
Incubation 1h30, 37°C | Incubation 1h30, 37°C | ||
| − | + | ===PCR=== | |
''by Coralie'' | ''by Coralie'' | ||
| − | + | * BBa_R0051 | |
We use the rehydrated plasmid from the iGEM plate 2014 | We use the rehydrated plasmid from the iGEM plate 2014 | ||
| Line 57: | Line 55: | ||
Keep it at 4°C | Keep it at 4°C | ||
| − | + | ===New culture=== | |
''by Seong Koo'' | ''by Seong Koo'' | ||
Observation of our plates: a lot of colony in each one. | Observation of our plates: a lot of colony in each one. | ||
New liquid culture of: | New liquid culture of: | ||
| − | * | + | * BBa_K1399005 |
| − | * | + | * BBa_K1399019 |
| − | * | + | * BBa_K1399023 |
| − | * Ligation product: | + | * Ligation product: BBa_J23101 + BBa_K115017 |
2x 5ml LB + 10μl Chloramphenicol + 1 bacterial colony. | 2x 5ml LB + 10μl Chloramphenicol + 1 bacterial colony. | ||
We incubate cultures at 37°C, ON. | We incubate cultures at 37°C, ON. | ||
| − | + | ===Plasmid extraction=== | |
''by Pauline'' | ''by Pauline'' | ||
| − | + | * BBa_R0051 | |
| − | * | + | * BBa_B0030 |
| − | * | + | |
Cultures from the 07/15/2015 | Cultures from the 07/15/2015 | ||
With the Nucleospin Kit from Macherey Nagel | With the Nucleospin Kit from Macherey Nagel | ||
| − | + | ===Digestion=== | |
''by Pauline'' | ''by Pauline'' | ||
| − | + | * BBa_R0051 (07/15/2015) | |
| − | * | + | * BBa_R0051 (07/16/2015) |
| − | * | + | * BBa_B0030 |
| − | * | + | * BBa_J23101 + BBa_I13504 |
| − | * | + | * BBa_J23106 + BBa_I13504 |
| − | * | + | * BBa_J23117 + BBa_I13504 |
| − | * | + | |
Reaction mix: | Reaction mix: | ||
| Line 97: | Line 93: | ||
* H2O: 15µL | * H2O: 15µL | ||
| − | + | ===Electrophoresis=== | |
''by Pauline'' | ''by Pauline'' | ||
Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET | Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET | ||
| − | Migration 0,06A 80V | + | Migration 0,06A 80V |
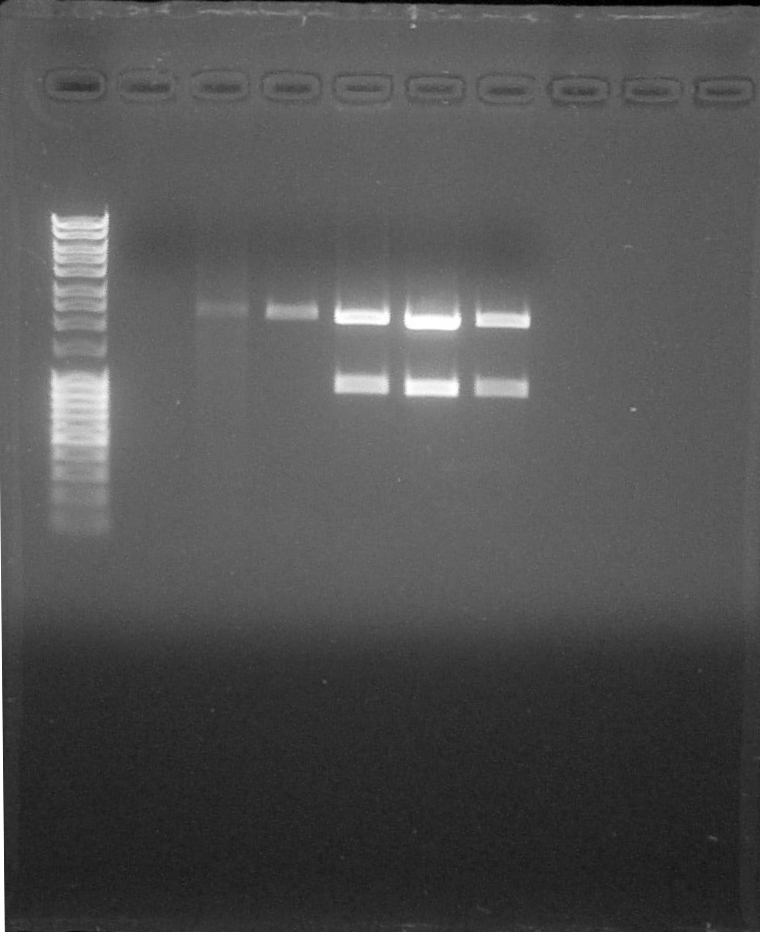
| − | + | [[File:ParisSaclay_16.07.15_-_digestion_vérif.jpg|300px|center]] | |
| − | ====Purification of | + | <html><i><p>Digestion verification, from left to right: 1. <a href="https://2015.igem.org/File:Paris_Saclay-Ladder.jpg" target="_blank">DNA Ladder</a>, 2. BBa_R0051 (15/07), 3. BBa_R0051 (16/07), 4. BBa_B0030#1, 5. BBa_J23101+I13504#1, 6. BBa_J23106+I13504#1, 7. BBa_J23117+I13504#1, 8. Empty, 9. Empty, 10. Empty</p></i></html> |
| + | ===Purification of BioBricks by electrophoresis=== | ||
''by Coralie'' | ''by Coralie'' | ||
| − | + | * BBa_S03518 | |
| − | * | + | * BBa_I13602 |
| − | * | + | |
Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET | Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET | ||
Migration 0,06A 80V | Migration 0,06A 80V | ||
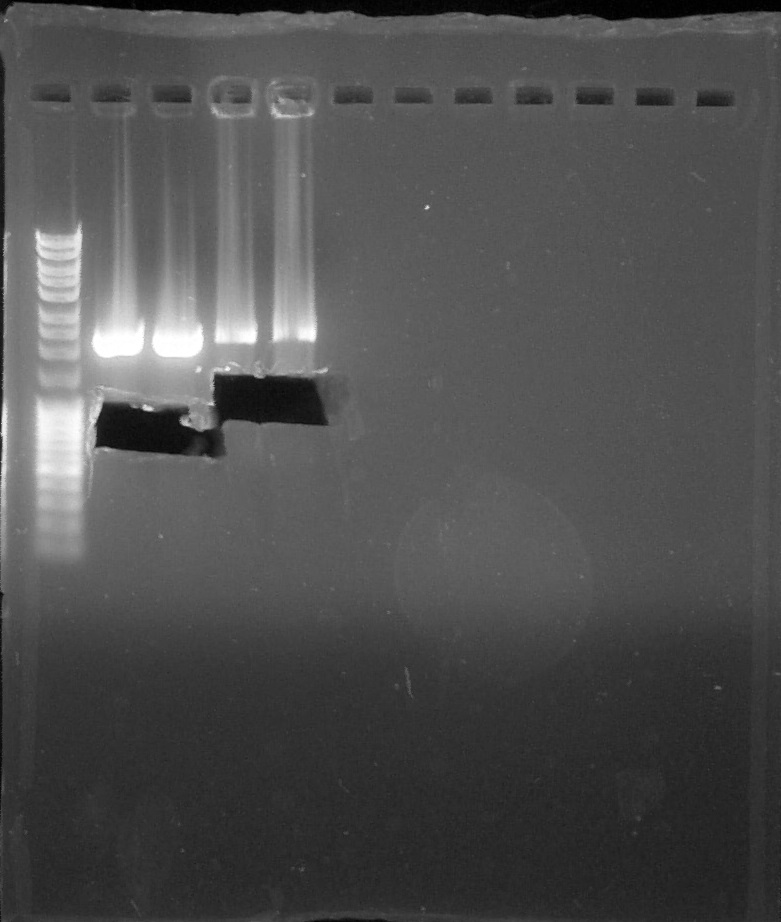
| + | [[File:ParisSaclay_16.07.2015_-_Purification.jpg|300px|center]] | ||
| + | <html><i><p>Purificatin verification, from left to right: 1. <a href="https://2015.igem.org/File:Paris_Saclay-Ladder.jpg" target="_blank">DNA Ladder</a>, 2. BBa_S03518#1, 3. BBa_S03518#2, 4. BBa_I13602#1, 5. BBa_I13602#2, 6. Empty, 7. Empty, 8. Empty, 9. Empty, 10. Empty, 11. Empty, 12. Empty</p></i></html> | ||
We cut interested bands with a scalpel. | We cut interested bands with a scalpel. | ||
| − | + | ===DNA Extraction and purification=== | |
''by Coralie'' | ''by Coralie'' | ||
| − | + | * BBa_S03518 #1 and #2 | |
| − | * | + | * BBa_B0015 #1 and #2 |
| − | * | + | * BBa_I13602 (x2) |
| − | * | + | * BBa_R0051 (from PCR) |
| − | * | + | |
We use the PCR Clean Up / Gel Extraction Kit from Macherey-Nagel | We use the PCR Clean Up / Gel Extraction Kit from Macherey-Nagel | ||
We elute DNA in 30 µL. | We elute DNA in 30 µL. | ||
| − | + | ===Quantification on Agarose Gel=== | |
''by Johan'' | ''by Johan'' | ||
| Line 131: | Line 128: | ||
We load 2µL of previous purified DNA with 2 µL of DNA Loading (6x) and 8 µL of H2O | We load 2µL of previous purified DNA with 2 µL of DNA Loading (6x) and 8 µL of H2O | ||
Migration 0,06A 80V | Migration 0,06A 80V | ||
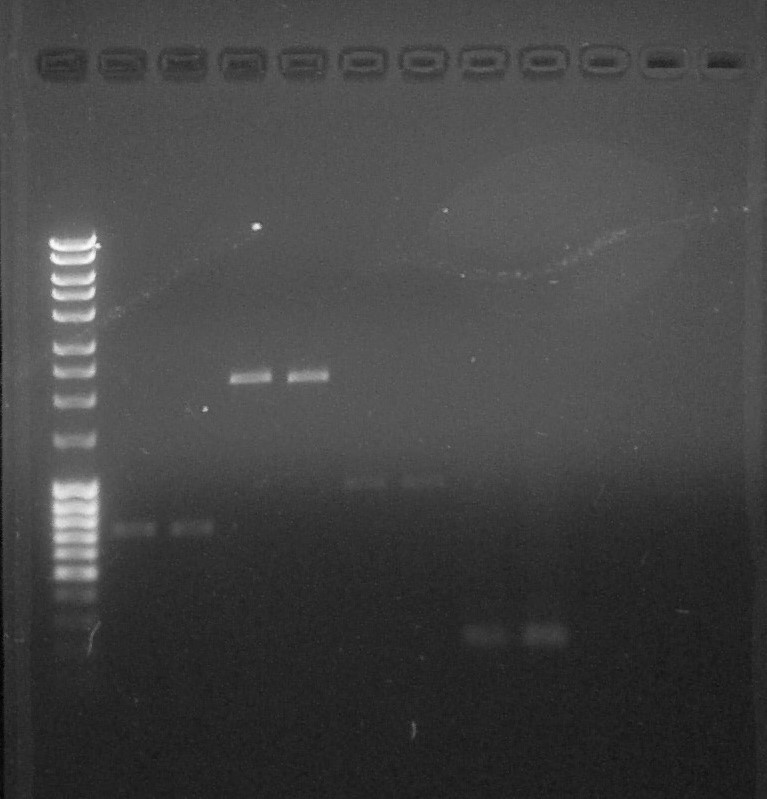
| − | + | [[File:ParisSaclay_16.07.15_-_quantif_pre-ligation.jpg|300px|center]] | |
| + | <html><i><p>Quantification, from left to right: 1. <a href="https://2015.igem.org/File:Paris_Saclay-Ladder.jpg" target="_blank">DNA Ladder</a>, 2. BBa_S03518#1, 3. BBa_S03518#2, 4. BBa_B0015#1, 5. BBa_B0015#2, 6. BBa_I13602#1, 7. BBa_I13602#2, 8. BBa_R0051#1, 9. BBa_BBa_R0051#2, 10. Empty, 11. Empty, 12. Empty</p></i></html> | ||
We can observe that the PCR of R0051 was effective. | We can observe that the PCR of R0051 was effective. | ||
We can quantify purified DNA: | We can quantify purified DNA: | ||
| − | * | + | * BBa_S03518 #1: 15 ng/µL |
| − | * | + | * BBa_S03518 #2: 15ng/µL |
| − | * | + | * BBa_B0015 #1: 10 ng/µL |
| − | * | + | * BBa_B0015 #2: 10 ng/µL |
| − | * | + | * BBa_I13602 (x2): 5ng/µL |
| − | * | + | * BBa_R0051: 10 ng/µL |
| − | + | ||
| − | + | ||
'''Members present:''' | '''Members present:''' | ||
* Instructors and advisors: Alice. | * Instructors and advisors: Alice. | ||
* Students: Johan, Seong Koo, Coralie, Pauline | * Students: Johan, Seong Koo, Coralie, Pauline | ||
| + | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 21:16, 18 September 2015
Contents
Thursday 16th July
Lab Work
Plasmid extraction
by Johan
- BBa_S03518
- BBa_B0015
Cultures from the 07/15/2015 With the Nucleospin Kit from Macherey Nagel
Digestion
by Coralie
- BBa_S03518
- BBa_B0015
- BBa_I13602
In each tube:
- Plasmid: 10µL
- Enzyme: 1µL of each enzyme
- Buffer FastDigest (10x): 2µL
- H2O: 6µL
Enzymes choice:
- BBa_S03518 #1: XbaI + PstI
- BBa_S03518 #2: SpeI + EcoRI
- BBa_B0015 #1: PstI + SpeI
- BBa_B0015 #2: XbaI + EcoRI
- BBa_I13602 (x2): XbaI + Pst I
Incubation 1h30, 37°C
PCR
by Coralie
- BBa_R0051
We use the rehydrated plasmid from the iGEM plate 2014
Reaction mix for 3 tubes:
- GC Buffer (5x): 30µL
- dNTP (10mM): 3µL
- Forward Primer (1/10): 7,5µL
- Reverse Primer (1/10): 7,5µL
- Template DNA R0051 (2014): 5µL
- DNA Pol Phusion: 1,5µL
- H2O: 97,5µL
We use 50µL of that mix in each tube
Cycle: Initiation: 98°C - 30seconds Cycle (34 repeats): 98°C - 10seconds / 65°C - 30seconds / 72°C - 20seconds Term.: 72°C - 5min Keep it at 4°C
New culture
by Seong Koo
Observation of our plates: a lot of colony in each one. New liquid culture of:
- BBa_K1399005
- BBa_K1399019
- BBa_K1399023
- Ligation product: BBa_J23101 + BBa_K115017
2x 5ml LB + 10μl Chloramphenicol + 1 bacterial colony. We incubate cultures at 37°C, ON.
Plasmid extraction
by Pauline
- BBa_R0051
- BBa_B0030
Cultures from the 07/15/2015 With the Nucleospin Kit from Macherey Nagel
Digestion
by Pauline
- BBa_R0051 (07/15/2015)
- BBa_R0051 (07/16/2015)
- BBa_B0030
- BBa_J23101 + BBa_I13504
- BBa_J23106 + BBa_I13504
- BBa_J23117 + BBa_I13504
Reaction mix:
- Plasmid: 2µL
- EcoRI: 0,5µL
- PstI: 0,5µL
- Buffer FastDigest (10x): 2µL
- H2O: 15µL
Electrophoresis
by Pauline
Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET Migration 0,06A 80V
Digestion verification, from left to right: 1. DNA Ladder, 2. BBa_R0051 (15/07), 3. BBa_R0051 (16/07), 4. BBa_B0030#1, 5. BBa_J23101+I13504#1, 6. BBa_J23106+I13504#1, 7. BBa_J23117+I13504#1, 8. Empty, 9. Empty, 10. Empty
Purification of BioBricks by electrophoresis
by Coralie
- BBa_S03518
- BBa_I13602
Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET Migration 0,06A 80V
Purificatin verification, from left to right: 1. DNA Ladder, 2. BBa_S03518#1, 3. BBa_S03518#2, 4. BBa_I13602#1, 5. BBa_I13602#2, 6. Empty, 7. Empty, 8. Empty, 9. Empty, 10. Empty, 11. Empty, 12. Empty
We cut interested bands with a scalpel.DNA Extraction and purification
by Coralie
- BBa_S03518 #1 and #2
- BBa_B0015 #1 and #2
- BBa_I13602 (x2)
- BBa_R0051 (from PCR)
We use the PCR Clean Up / Gel Extraction Kit from Macherey-Nagel We elute DNA in 30 µL.
Quantification on Agarose Gel
by Johan
Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET We load 2µL of previous purified DNA with 2 µL of DNA Loading (6x) and 8 µL of H2O Migration 0,06A 80V
Quantification, from left to right: 1. DNA Ladder, 2. BBa_S03518#1, 3. BBa_S03518#2, 4. BBa_B0015#1, 5. BBa_B0015#2, 6. BBa_I13602#1, 7. BBa_I13602#2, 8. BBa_R0051#1, 9. BBa_BBa_R0051#2, 10. Empty, 11. Empty, 12. Empty
We can observe that the PCR of R0051 was effective. We can quantify purified DNA:- BBa_S03518 #1: 15 ng/µL
- BBa_S03518 #2: 15ng/µL
- BBa_B0015 #1: 10 ng/µL
- BBa_B0015 #2: 10 ng/µL
- BBa_I13602 (x2): 5ng/µL
- BBa_R0051: 10 ng/µL
Members present:
- Instructors and advisors: Alice.
- Students: Johan, Seong Koo, Coralie, Pauline