Difference between revisions of "Team:Chalmers-Gothenburg/SafetySwitch"
| Line 1: | Line 1: | ||
| − | + | {{Chalmers-Gothenburg}} | |
<h2>Safety Switch</h2> | <h2>Safety Switch</h2> | ||
| − | <h3 | + | <h3>Defining the model<h3> |
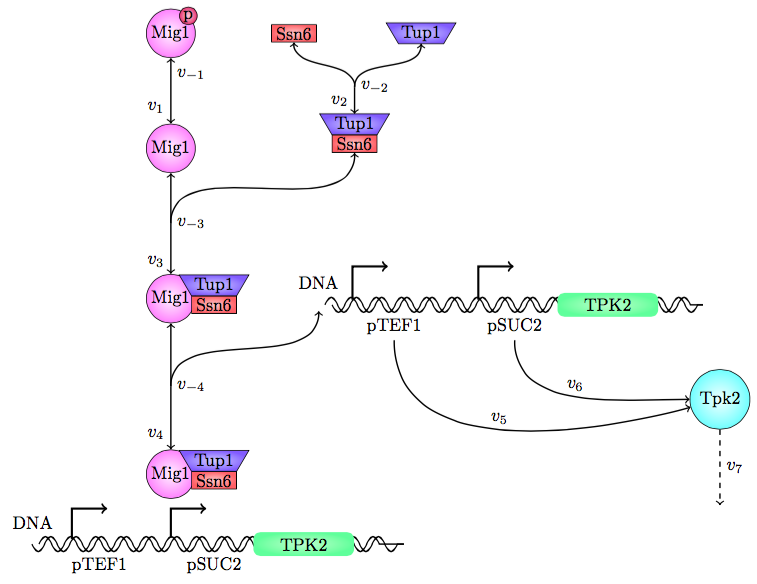
| − | < | + | <p>The safety switch is based on using the regulation of the SUC2 gene to express a potential harmful kinase domain Tpk2. A well know, always on, overexpression promotor pTEF1 was used to amplify the protein expression while the first promotor, pSUC2 is not down regulated. SUC2 is downregulated by the binding if a Zink finger containing protein Mig1 which needs to be in a complex with the Co repressor SSN6/Tup1 to be active as a repressor. </p> |
| − | < | + | <p>There have been quite a lot of modeling done one the Mig1 activity based on sugar abundance in the media. 2009, Frey et al managed to create a model with predictable predictability. [1] The model used a heavy side function describe the activity, Mig1 which is a of the Zinc finger containing protein involved in repressing the SUC2 gene. The heavy side functions on or of state was based on the glucose level in the media. Due to the good predictability of the Mig1 model, the first corner stone of the model was set. Non-associated Ssn+6 and Tup1 was assumed to be relevant for the expression and was therefore included in the model. The repression mechanism used in the model was a physical mechanism, when the repressor is associated to the promotor, no gene expression is active. And the assumption that a repressor free promoter leads to a linear gene expression was made.</p> |
[[File:Chalmersgothenburgssmodel1.png|700px]] | [[File:Chalmersgothenburgssmodel1.png|700px]] | ||
| − | <p>Figure 1<p> | + | <p class="figuretext">Figure 1, the modeled pathway in the Safety switch model.</p> |
| − | <p>The | + | <p>The reactions were defined as:</p> |
| − | |||
[[File:Chalmersgothenburgssmodel2.png|300px]] | [[File:Chalmersgothenburgssmodel2.png|300px]] | ||
| − | Yielding in the following reaction equations. | + | |
| + | <p>Yielding in the following reaction equations.</p> | ||
[[File:Chalmersgothenburgssmodel3.png|200px]] | [[File:Chalmersgothenburgssmodel3.png|200px]] | ||
| − | + | <h3>Parameters<h3> | |
| − | <h3 | + | |
Parameters k1, k-1, k2 and k-2 were found in the literature. However, the reaction constant for Reaction 3 were only found as a disassociation constants. An arbitrary number was set as one of the reaction, 2 × 10^2 m −1 s−1 and the other parameter was calculated based on the disassociation constants and the arbitrary number. | Parameters k1, k-1, k2 and k-2 were found in the literature. However, the reaction constant for Reaction 3 were only found as a disassociation constants. An arbitrary number was set as one of the reaction, 2 × 10^2 m −1 s−1 and the other parameter was calculated based on the disassociation constants and the arbitrary number. | ||
For reaction 4 two a disassociation constants were found due to the Mig1 having zinc fingers and both binding to the DNA strand, by random the larger value were chosen [5]. We looked into a potential ways of modelling the two zinc finger/ DNA association. When the first zinc finger is bounded to DNA, while in that state the possible volume for the second finger is highly decreased and the local concentration will be increased. However we had no way of determining the possible volume for the second association. The idea was therefore dropped. | For reaction 4 two a disassociation constants were found due to the Mig1 having zinc fingers and both binding to the DNA strand, by random the larger value were chosen [5]. We looked into a potential ways of modelling the two zinc finger/ DNA association. When the first zinc finger is bounded to DNA, while in that state the possible volume for the second finger is highly decreased and the local concentration will be increased. However we had no way of determining the possible volume for the second association. The idea was therefore dropped. | ||
| Line 49: | Line 48: | ||
4. Brinker, A., Scheufler, C., Von der Mülbe, F., Fleckenstein, B., Herrmann, C., Jung, G., ... & Hartl, F. U. (2002). Ligand discrimination by TPR domains relevance and selectivity of eevd-recognition in Hsp70· Hop· Hsp90 complexes. Journal of Biological Chemistry, 277(22), 19265-19275. | 4. Brinker, A., Scheufler, C., Von der Mülbe, F., Fleckenstein, B., Herrmann, C., Jung, G., ... & Hartl, F. U. (2002). Ligand discrimination by TPR domains relevance and selectivity of eevd-recognition in Hsp70· Hop· Hsp90 complexes. Journal of Biological Chemistry, 277(22), 19265-19275. | ||
5 Needham, P. G., & Trumbly, R. J. (2006). In vitro characterization of the Mig1 repressor from Saccharomyces cerevisiae reveals evidence for monomeric and higher molecular weight forms. Yeast, 23(16), 1151-1166. | 5 Needham, P. G., & Trumbly, R. J. (2006). In vitro characterization of the Mig1 repressor from Saccharomyces cerevisiae reveals evidence for monomeric and higher molecular weight forms. Yeast, 23(16), 1151-1166. | ||
| − | |||
6. Williams, T. C., Espinosa, M. I., Nielsen, L. K., & Vickers, C. E. (2015). Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae. Microbial cell factories, 14(1), 43. | 6. Williams, T. C., Espinosa, M. I., Nielsen, L. K., & Vickers, C. E. (2015). Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae. Microbial cell factories, 14(1), 43. | ||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
Revision as of 23:41, 18 September 2015