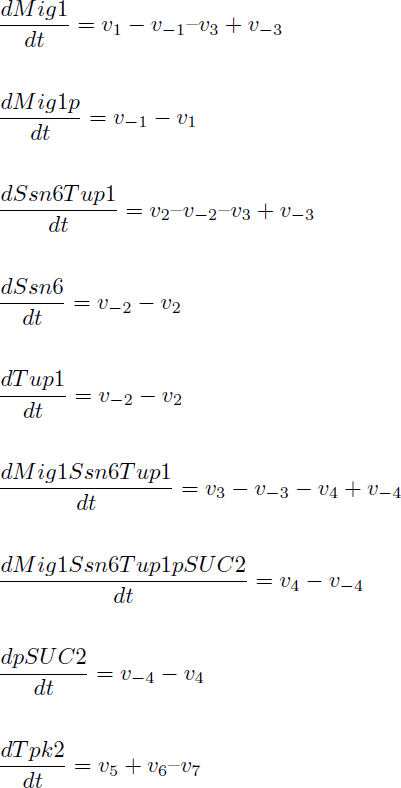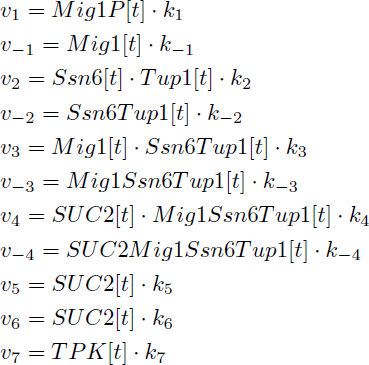Difference between revisions of "Team:Chalmers-Gothenburg/SafetySwitch"
| Line 100: | Line 100: | ||
<p>We found no absolute data on the amount of proteins produced from the specific promoters, we therefore lumped all the reactions and gave it an arbitrary magnitude to get a feeling of the relative expression of the different systems and conditions. However due to this fact there is no quantitative predictability of the resulting protein levels, only relative. The literature study also indicated that the promoter had similar relative strength while SUC2 was not down regulated [6].</p> | <p>We found no absolute data on the amount of proteins produced from the specific promoters, we therefore lumped all the reactions and gave it an arbitrary magnitude to get a feeling of the relative expression of the different systems and conditions. However due to this fact there is no quantitative predictability of the resulting protein levels, only relative. The literature study also indicated that the promoter had similar relative strength while SUC2 was not down regulated [6].</p> | ||
| + | |||
| + | |||
[[File:ChalmersGothenburgKTABLE.png|700px]] | [[File:ChalmersGothenburgKTABLE.png|700px]] | ||
<p class="figuretext">Table 1, The rates of the reaction constants and their sources.</p> | <p class="figuretext">Table 1, The rates of the reaction constants and their sources.</p> | ||
| + | |||
| + | |||
| + | |||
<table style="width:100%; border-collapse: collapse"> | <table style="width:100%; border-collapse: collapse"> | ||
| Line 129: | Line 134: | ||
<td>2220 molecules/cell</td> | <td>2220 molecules/cell</td> | ||
</tr> | </tr> | ||
| + | <tr> | ||
| + | <td>Rest</td> | ||
| + | <td>0 molecules/cell</td> | ||
| + | </tr> | ||
| + | |||
</table> | </table> | ||
| + | <p class="figuretext">Table 2, The boundary conditions for the model [7] .</p> | ||
| + | |||
<h3> References </h3> | <h3> References </h3> | ||
| Line 139: | Line 151: | ||
<p>[6] Williams, T. C., Espinosa, M. I., Nielsen, L. K., & Vickers, C. E. (2015). Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae. Microbial cell factories, 14(1), 43.</p> | <p>[6] Williams, T. C., Espinosa, M. I., Nielsen, L. K., & Vickers, C. E. (2015). Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae. Microbial cell factories, 14(1), 43.</p> | ||
| + | <p> [7] Chong, Y. T., Koh, J. L., Friesen, H., Duffy, K., Cox, M. J., Moses, A., ... & Andrews, B. J. (2015). Yeast proteome dynamics from single cell imaging and automated analysis. Cell, 161(6), 1413-1424. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 03:17, 19 September 2015


Safety Switch
SUC
Figure 1. The concentration, in Mol, of active promoters over time in the presence of glucose.
Figure 2. The concentration, in Mol, of active promoters over time in the absence of glucose.
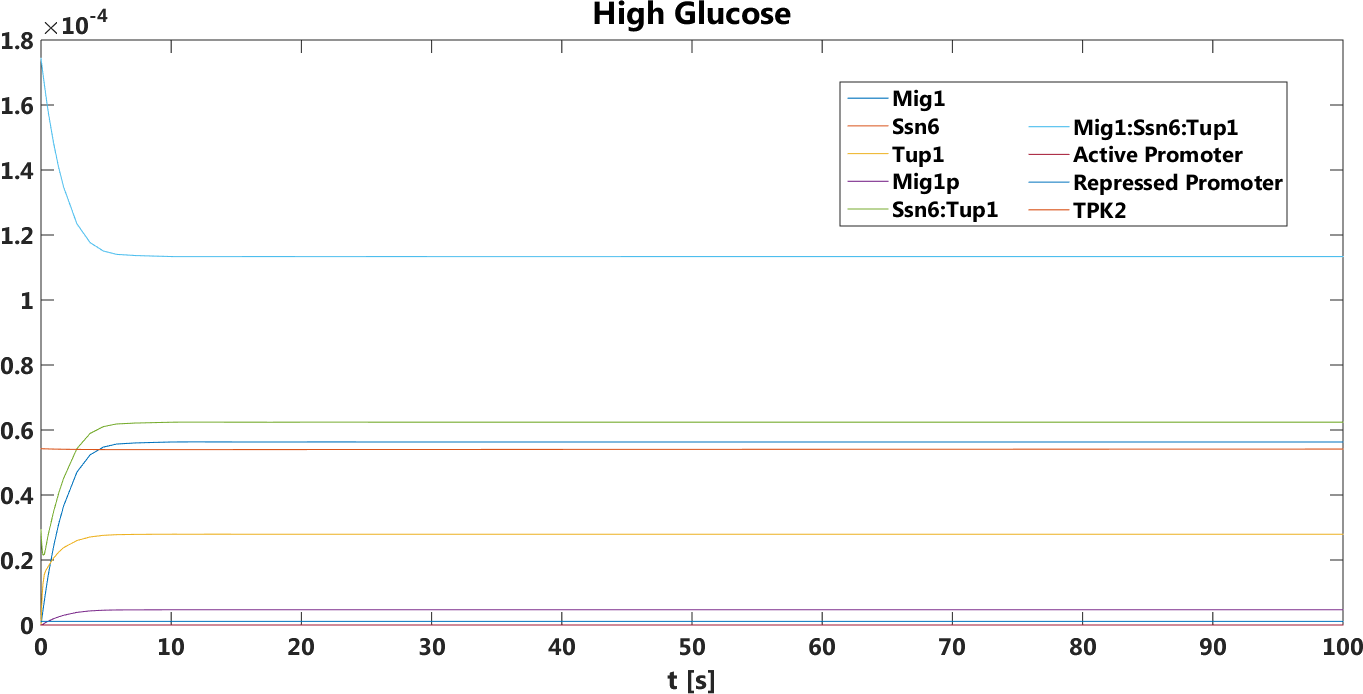
Figure 3. The concentration, in Mol, of all components over time in the presence of glucose.
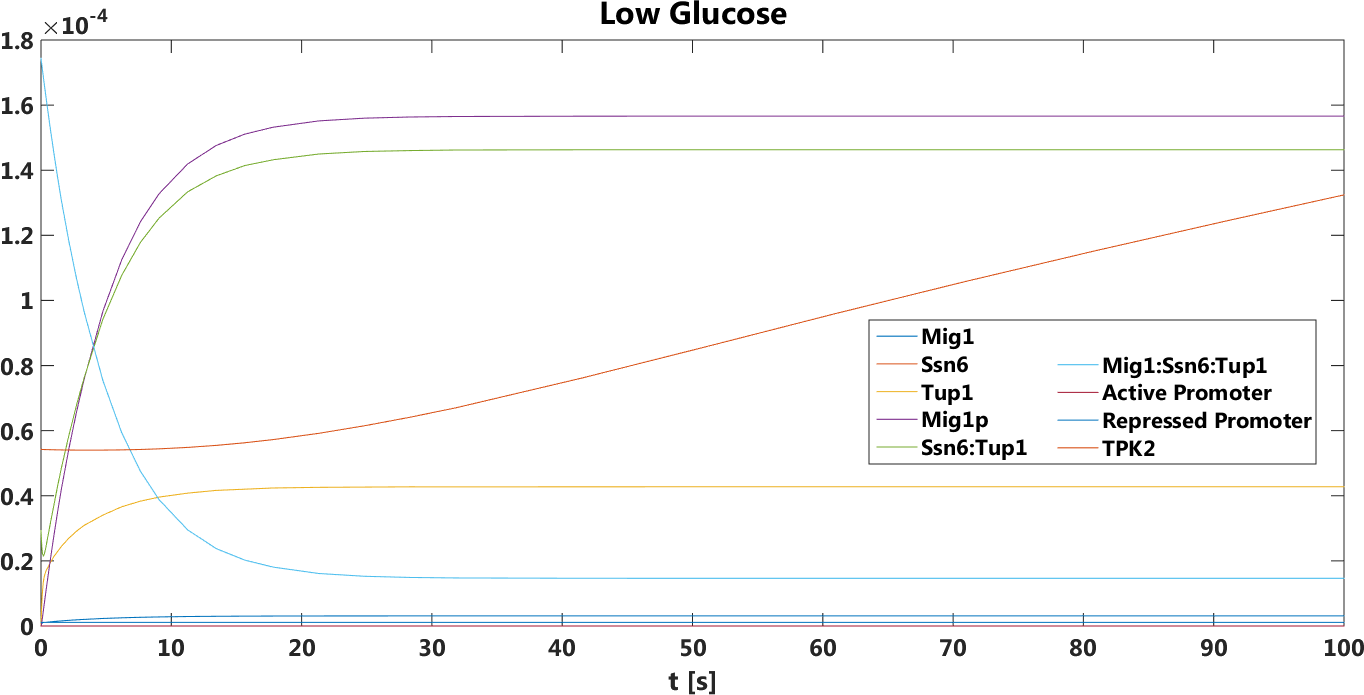
Figure 4. The concentration, in Mol, of all components over time in the absence of glucose.
Figure 5. The concentration, in Mol, of Mig related components over time in the presence of glucose.
Figure 6. The concentration, in Mol, of Mig related components over time in the absence of glucose.
Figure 7. The concentration, in Mol, of repressed promoters over time in the presence of glucose.
Figure 8. The concentration, in Mol, of repressed promoters over time in the absence of glucose.
Figure 9. The concentration, in Mol, of TPK2 over time in the presence of glucose.
Figure 10. The concentration, in Mol, of TPK2 over time in the absence of glucose.
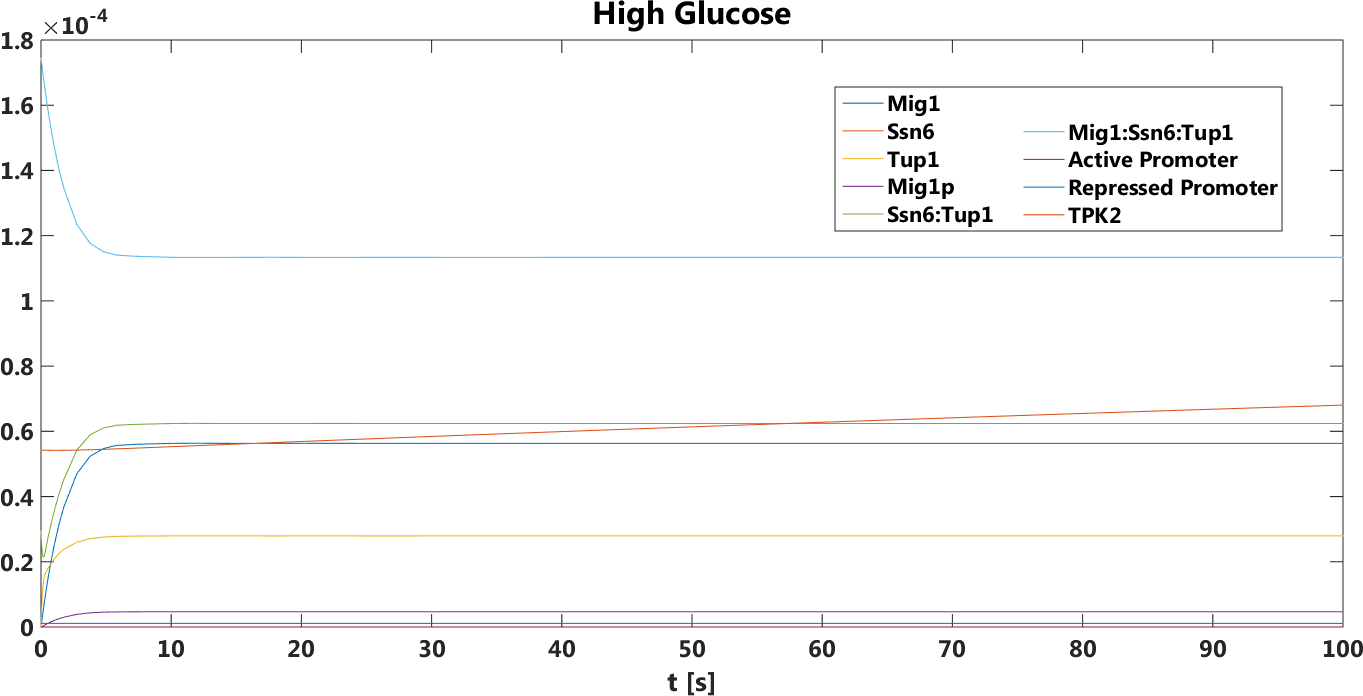
Figure 11. The concentration, in Mol, of Tuf and Ssn6 related components over time in the presence of glucose.
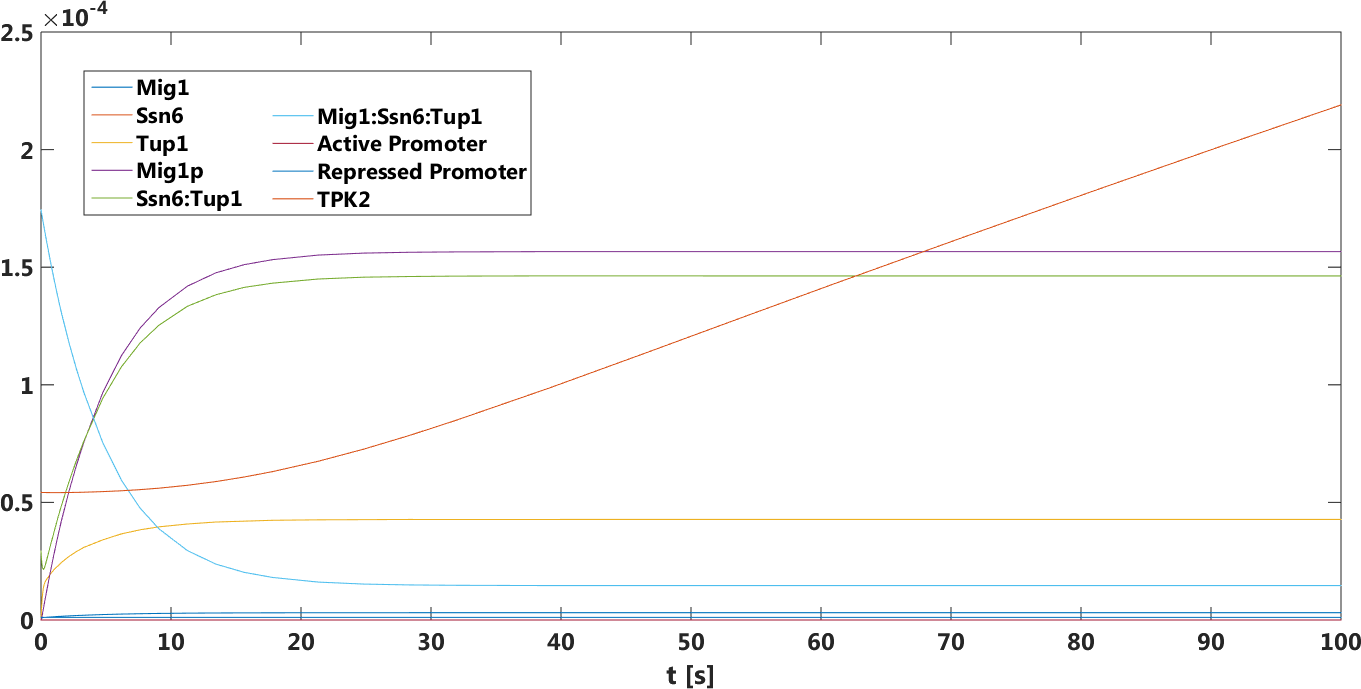
Figure 12. The concentration, in Mol, of Tuf and Ssn6 related components over time in the absence of glucose.
TEFSUC
Figure 13. The concentration, in Mol, of active promoters over time in the presence of glucose.
Figure 14. The concentration, in Mol, of active promoters over time in the absence of glucose.
Figure 15. The concentration, in Mol, of all components over time in the presence of glucose.
Figure 16. The concentration, in Mol, of all components over time in the absence of glucose.
Figure 17. The concentration, in Mol, of Mig related components over time in the presence of glucose.
Figure 18. The concentration, in Mol, of Mig related components over time in the absence of glucose.
Figure 19. The concentration, in Mol, of repressed promoters over time in the presence of glucose.
Figure 20. The concentration, in Mol, of repressed promoters over time in the absence of glucose.
Figure 21. The concentration, in Mol, of TPK2 over time in the presence of glucose.
Figure 22. The concentration, in Mol, of TPK2 over time in the absence of glucose.
Figure 23. The concentration, in Mol, of Tuf and Ssn6 related components over time in the presence of glucose.
Figure 24. The concentration, in Mol, of Tuf and Ssn6 related components over time in the absence of glucose.
Defining the model
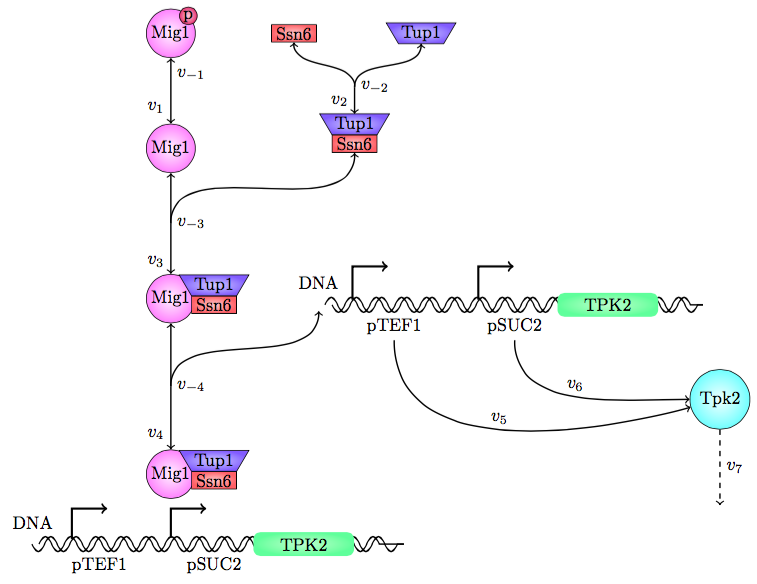
The safety switch is based on using the regulation of the SUC2 gene to express a potential harmful kinase domain Tpk2. A well know, always on, overexpression promotor pTEF1 was used to amplify the protein expression while the first promotor, pSUC2 is not down regulated. SUC2 is downregulated by the binding if a Zink finger containing protein Mig1 which needs to be in a complex with the Co repressor SSN6/Tup1 to be active as a repressor.
There have been quite a lot of modeling done one the Mig1 activity based on sugar abundance in the media. 2009, Frey et al managed to create a model with predictable predictability. [1] The model used a heavy side function describe the activity, Mig1 which is a of the Zinc finger containing protein involved in repressing the SUC2 gene. The heavy side functions on or of state was based on the glucose level in the media. Due to the good predictability of the Mig1 model, the first corner stone of the model was set. Non-associated Ssn+6 and Tup1 was assumed to be relevant for the expression and was therefore included in the model. The repression mechanism used in the model was a physical mechanism, when the repressor is associated to the promotor, no gene expression is active. And the assumption that a repressor free promoter leads to a linear gene expression was made.
Figure 25, the modeled pathway in the Safety switch model.
The reactions were defined as:
Yielding in the following reaction equations.
Parameters
Parameters k1, k-1, k2 and k-2 were found in the literature. However, the reaction constant for Reaction 3 were only found as a disassociation constant. An arbitrary number was set as one of the reaction, 2×10^2 /(m*s)and the other parameter was calculated based on the disassociation constant and the arbitrary number. For reaction 4, two disassociation constants were found due to the Mig1 having zinc fingers and both binding to the DNA strand, the larger value was chosen randomly [5]. We looked into a potential way of modelling the two zinc finger/DNA association, specifically when the first zinc finger is bound to DNA. While in that state the possible volume for the second finger is highly decreased and the local concentration will be increased. However we had no way of determining the possible volume for the second association. The idea was therefore dropped.
We found no absolute data on the amount of proteins produced from the specific promoters, we therefore lumped all the reactions and gave it an arbitrary magnitude to get a feeling of the relative expression of the different systems and conditions. However due to this fact there is no quantitative predictability of the resulting protein levels, only relative. The literature study also indicated that the promoter had similar relative strength while SUC2 was not down regulated [6].
Table 1, The rates of the reaction constants and their sources.
| [Ssn6] | 3890 molecules/cell |
| [Tup1] | 5840 molecules/cell |
| [Mig1] | 830 molecules/cell |
| [pSUC2] | 2 molecules/cell |
| [PTEF1] | 2 molecules/cell |
| [TPK2] | 2220 molecules/cell |
| Rest | 0 molecules/cell |
Table 2, The boundary conditions for the model [7] .
References
[1] Frey, S., Sott, K., Smedh, M., Millat, T., Dahl, P., Wolkenhauer, O., & Goksör, M. (2011). A mathematical analysis of nuclear intensity dynamics for Mig1-GFP under consideration of bleaching effects and background noise in Saccharomyces cerevisiae. Molecular BioSystems, 7(1), 215-223
[2] García‐Salcedo, R., Lubitz, T., Beltran, G., Elbing, K., Tian, Y., Frey, S., ... & Hohmann, S. (2014). Glucose de‐repression by yeast AMP‐activated protein kinase SNF1 is controlled via at least two independent steps. Febs Journal, 281(7), 1901-1917.
[3] Jabet, C., Sprague, E. R., VanDemark, A. P., & Wolberger, C. (2000). Characterization of the N-terminal Domain of the Yeast Transcriptional Repressor Tup1 PROPOSAL FOR AN ASSOCIATION MODEL OF THE REPRESSOR COMPLEX Tup1· Ssn6. Journal of Biological Chemistry, 275(12), 9011-9018.
[4] Brinker, A., Scheufler, C., Von der Mülbe, F., Fleckenstein, B., Herrmann, C., Jung, G., ... & Hartl, F. U. (2002). Ligand discrimination by TPR domains relevance and selectivity of eevd-recognition in Hsp70· Hop· Hsp90 complexes. Journal of Biological Chemistry, 277(22), 19265-19275.
[5] Needham, P. G., & Trumbly, R. J. (2006). In vitro characterization of the Mig1 repressor from Saccharomyces cerevisiae reveals evidence for monomeric and higher molecular weight forms. Yeast, 23(16), 1151-1166.
[6] Williams, T. C., Espinosa, M. I., Nielsen, L. K., & Vickers, C. E. (2015). Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae. Microbial cell factories, 14(1), 43.
[7] Chong, Y. T., Koh, J. L., Friesen, H., Duffy, K., Cox, M. J., Moses, A., ... & Andrews, B. J. (2015). Yeast proteome dynamics from single cell imaging and automated analysis. Cell, 161(6), 1413-1424.