Difference between revisions of "Team:Exeter/Interlab"
| (115 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| + | <div class="container"> | ||
| − | <script src="https://2015.igem.org/Team:Exeter/ajax | + | <script src="https://2015.igem.org/Team:Exeter/ajax-googleapis-com/ajax/libs/jquery/1-8-2/jquery-min?action=raw&ctype=text/javascript"></script> |
| − | <script src="https://2015.igem.org/Team:Exeter/js/jquery | + | <script src="https://2015.igem.org/Team:Exeter/js/jquery-bxslider-min?action=raw&ctype=text/javascript"></script> |
| − | + | ||
| − | + | ||
| + | <link href="https://2015.igem.org/Team:Exeter/lib/jquery-bxslider?action=raw&ctype=text/css" rel="stylesheet"> | ||
<script> | <script> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
//Slider JS | //Slider JS | ||
$(document).ready(function(){ | $(document).ready(function(){ | ||
| − | $('.pro, .ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data, .proto').show('fast'); | + | //$('.pro, .ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data, .proto').show('fast'); |
var slider = $('.slider500').bxSlider({ | var slider = $('.slider500').bxSlider({ | ||
| Line 30: | Line 24: | ||
captions: true, | captions: true, | ||
adaptiveHeight: true, | adaptiveHeight: true, | ||
| − | preloadImages: 'all' | + | preloadImages: 'all', |
| + | pager: false | ||
}); | }); | ||
| Line 40: | Line 35: | ||
captions: true, | captions: true, | ||
adaptiveHeight: true, | adaptiveHeight: true, | ||
| − | preloadImages: 'all' | + | preloadImages: 'all', |
| + | pager: false | ||
}); | }); | ||
| Line 50: | Line 46: | ||
captions: true, | captions: true, | ||
adaptiveHeight: true, | adaptiveHeight: true, | ||
| − | preloadImages: 'all' | + | preloadImages: 'all', |
| + | pager: false | ||
}); | }); | ||
| + | $('.sliderstatic400').bxSlider({ | ||
| + | slideWidth: 400, | ||
| + | minSlides: 1, | ||
| + | maxSlides: 1, | ||
| + | slideMargin: 0, | ||
| + | captions: true, | ||
| + | adaptiveHeight: true, | ||
| + | preloadImages: 'all', | ||
| + | controls: false, | ||
| + | touchEnabled: false, | ||
| + | pager: false | ||
| + | }); | ||
| − | $('. | + | $('.sliderstatic300').bxSlider({ |
| + | slideWidth: 300, | ||
| + | minSlides: 1, | ||
| + | maxSlides: 1, | ||
| + | slideMargin: 0, | ||
| + | captions: true, | ||
| + | adaptiveHeight: true, | ||
| + | preloadImages: 'all', | ||
| + | controls: false, | ||
| + | touchEnabled: false, | ||
| + | pager: false | ||
| + | }); | ||
| − | }); | + | $('.sliderstatic500').bxSlider({ |
| + | slideWidth: 500, | ||
| + | minSlides: 1, | ||
| + | maxSlides: 1, | ||
| + | slideMargin: 0, | ||
| + | captions: true, | ||
| + | adaptiveHeight: true, | ||
| + | preloadImages: 'all', | ||
| + | controls: false, | ||
| + | touchEnabled: false, | ||
| + | pager: false | ||
| + | }); | ||
| + | //$('.Loading').hide('slow'); | ||
| + | //$('.ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data, .proto').hide('slow'); | ||
| + | }); | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | </script> | ||
| − | |||
| − | |||
<style> | <style> | ||
| Line 103: | Line 120: | ||
padding-left: 20px; | padding-left: 20px; | ||
} | } | ||
| − | + | /* | |
| − | + | .level_3 a { | |
| − | + | width:40px; | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | width: | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
} | } | ||
| − | + | li.level_3 { | |
| + | min-width:70px; | ||
| + | height:20px; | ||
| + | } | ||
| + | */ | ||
</style> | </style> | ||
| Line 130: | Line 141: | ||
<h1>Interlab Study</h1> | <h1>Interlab Study</h1> | ||
| − | <!--Begin Level 3 Buttons | + | <!--Begin Level 3 Buttons |
| − | < | + | <center id="topmenu"> |
| − | <ul class=" | + | <ul class="pagination"> |
| − | <li | + | <li> |
<a onclick="$('.ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data, .proto').hide('slow');$('.pro').toggle('slow');"> | <a onclick="$('.ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data, .proto').hide('slow');$('.pro').toggle('slow');"> | ||
| − | + | 0 | |
</a> | </a> | ||
| − | </li> | + | </li> |
| − | <li | + | <li><a onclick="$('.pro, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data, .proto').hide('slow');$('.ch1').toggle('slow');"> |
| − | + | 1 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li><a onclick="$('.pro, .ch1, .ch3, .ch4, .ch5, .ch6, .ch7, .data, .proto').hide('slow');$('.ch2').toggle('slow')"> |
| − | + | 2 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li><a onclick="$('.pro, .ch1, .ch2, .ch4, .ch5, .ch6, .ch7, .data').hide('slow');$('.ch3').toggle('slow')"> |
| − | + | 3 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li><a onclick="$('.pro, .ch1, .ch2, .ch3, .ch5, .ch6, .ch7, .data').hide('slow');$('.ch4').toggle('slow')"> |
| − | + | 4 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li><a onclick="$('.pro, .ch1, .ch2, .ch3, .ch4, .ch6, .ch7, .data, .proto').hide('slow');$('.ch5').toggle('slow')"> |
| − | + | 5 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li><a onclick="$('.pro, .ch1, .ch2, .ch3, .ch4, .ch5, .ch7, .data, .proto').hide('slow');$('.ch6').toggle('slow')"> |
| − | + | 6 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li><a onclick="$('.pro, .ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .data').hide('slow');$('.ch7').toggle('slow')"> |
| − | + | 7 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li><a onclick="$('.pro, .ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .proto').hide('slow');$('.data').toggle('slow')"> |
| − | + | 8 | |
</a> | </a> | ||
</li> | </li> | ||
| − | <li | + | <li> |
<a onclick="$('.pro, .ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data').hide('slow');$('.proto').toggle('slow')"> | <a onclick="$('.pro, .ch1, .ch2, .ch3, .ch4, .ch5, .ch6, .ch7, .data').hide('slow');$('.proto').toggle('slow')"> | ||
| − | + | 9 | |
</a> | </a> | ||
| − | </li> | + | </li> |
| − | + | ||
</ul> | </ul> | ||
| − | </ | + | </center> |
<!--End Level 3 Buttons--> | <!--End Level 3 Buttons--> | ||
| − | |||
| − | |||
<!--Begin Prologue--> | <!--Begin Prologue--> | ||
| Line 285: | Line 293: | ||
Determined to contribute to the field of synthetic biology, we decided to participate in the Second International Interlab Measurement Study. While determining fluorescence levels for the three Interlab devices, we hoped to gain some experience in the lab before starting our iGEM project, Ribonostics. | Determined to contribute to the field of synthetic biology, we decided to participate in the Second International Interlab Measurement Study. While determining fluorescence levels for the three Interlab devices, we hoped to gain some experience in the lab before starting our iGEM project, Ribonostics. | ||
| − | We aimed to build and measure the fluorescence of three BioBrick devices: | + | We aimed to build and measure the fluorescence of three following BioBrick devices into pSB1C3 plasmid backbone: |
<ol> | <ol> | ||
<li>J23101 + I13504 (D1)</li> | <li>J23101 + I13504 (D1)</li> | ||
| Line 296: | Line 304: | ||
</ol> | </ol> | ||
| − | + | ||
</p> | </p> | ||
| Line 302: | Line 310: | ||
<p> | <p> | ||
| − | In the first week of our iGEM induction, we started out by hydrating DNA from the kit, after which we followed the standard protocols for BioBrick assembly. The first step was transforming | + | In the first week of our iGEM induction, we started out by re-hydrating the required DNA from the kit, after which we followed the standard protocols for BioBrick assembly. The first step of this process was transforming the BioBrick DNA into competent DH5α cells (from New England Biolabs, NEB), before growing them up on agar plates containing the correct antibiotic (ampicillin for I13504, chloramphenicol in the promoters). We then prepared overnight cultures. After transforming, we performed our first miniprep to obtain the DNA needed for further experiments, and checked the DNA concentration using Qubit. We then used values obtained from this to calculate the volumes of all of components of the digestion step. After the digestion, we ran a part of each sample on a gel to see if the insert and vector were the correct size. The ligation was the last step; after this, we plated our samples and incubated them overnight. We hoped that the next morning, there would be glowing green colonies on our plates. |
| − | Protocols of these experiments can be found | + | Protocols of these experiments can be found <a href="https://2015.igem.org/Team:Exeter/Experiments" >here </a>: transformation, miniprep, Qubit, digestion, ligation. |
</p> | </p> | ||
<p> | <p> | ||
| − | The next day, we came into the lab, hoping to see colonies so green they would be blinding. After viewing the plates under | + | The next day, we came into the lab, hoping to see colonies so green they would be blinding. After viewing the plates under UV light to visualise the green fluorescent protein (GFP) in our constructs, we understood that the statement ‘biology sometimes doesn’t work’ is entirely accurate. There were no glowing colonies on our plates. However, this moment of truth inspired us to diagnose what was wrong with our devices, and give the Interlab Study another try in the following week. |
</p> | </p> | ||
| Line 331: | Line 339: | ||
<p> | <p> | ||
| − | After a weekend of thought, we came into the lab, determined to get glowing colonies by the end of the week. The answer to why our | + | After a weekend of thought, we came into the lab, determined to get glowing colonies by the end of the week. The answer to why our interlab study failed the previous week was simple: the promoters we were using were not BioBrick assembly compatible – they did not join, because they could not join. |
</p> | </p> | ||
<p> | <p> | ||
| − | We looked at our second attempt at the | + | We looked at our second attempt at the interlab study with optimism – we had some practice in the lab, and we understood more clearly what it was we were actually trying to do. This time, we made and used TOP10 competent cells. Again, we followed the same flowchart of experiments, using the correct BioBricks (J23101, J23106, J23117). |
</p> | </p> | ||
| Line 361: | Line 369: | ||
<p> | <p> | ||
| − | By now, you may have guessed that our team is not the luckiest when it comes to glowing. If so, you guessed right – we did not have any glowing colonies. | + | By now, you may have guessed that our team is not the luckiest when it comes to glowing. If so, you guessed right – we did not have any glowing colonies. |
</p> | </p> | ||
<p> | <p> | ||
| − | We were baffled, puzzled, surprised – but not broken. Once again, we marched into the lab, determined to see fluorescent colonies. We double (and triple) checked that we were using the correct BioBricks, and enzymes during the digestion step. We also thought carefully about what we could change in order to get our Interlab Study to work. During our third attempt, we followed a protocol for a traditional digestion instead of the digestion protocol we had used before. We also altered the transformation protocol slightly. After running our digest on the gel, we performed a gel extraction – this way we would be sure that we were using vector and insert fragments of the correct sizes. Amongst the things we added and changed were also: control plates of just our competent cells, and using 1 µl of | + | We were baffled, puzzled, surprised – but not broken. Once again, we marched into the lab, determined to see fluorescent colonies. We double (and triple) checked that we were using the correct BioBricks, and enzymes during the digestion step. We also thought carefully about what we could change in order to get our Interlab Study to work. During our third attempt, we followed a protocol for a traditional digestion instead of the digestion protocol we had used before. We also altered the transformation protocol slightly. After running our digest on the gel, we performed a gel extraction – this way we would be sure that we were using vector and insert fragments of the correct sizes. Amongst the things we added and changed were also: control plates of just our competent cells, and using 1 µl of fresh T4 ligase instead of 0.5 µl. Again, we followed the Interlab flowchart of procedures. Armed with our transformed Interlab samples and plenty of controls, we saw no reason for our colonies not to glow the next morning. |
</p> | </p> | ||
| Line 384: | Line 392: | ||
<p> | <p> | ||
| − | On the rainy Friday morning, we climbed the stairs to the fourth floor, where our lab was located. We had great – but realistic – expectations for our colonies. Once we saw them, we knew the likely result of our third attempt at the Interlab Study. The | + | On the rainy Friday morning, we climbed the stairs to the fourth floor, where our lab was located. We had great – but realistic – expectations for our colonies. Once we saw them, we knew the likely result of our third attempt at the Interlab Study. The UV light confirmed our suspicions – we did not have any green glowing colonies. |
We had a discussion about what could be going wrong with the Interlab process, and came up with three hypotheses: | We had a discussion about what could be going wrong with the Interlab process, and came up with three hypotheses: | ||
| Line 390: | Line 398: | ||
<ul> | <ul> | ||
| − | <li>During our ligation, we used a 1:1 ratio of insert to vector – perhaps using a 3:1 or 1:3 ratio would be better.</li> | + | <li>During our ligation, we used a 1:1 ratio of insert to vector – perhaps using a 3:1 or 1:3 ratio would be better. </li> |
| − | <li>We always incubated our ligation overnight in the fridge – maybe incubating at room temperature would work more efficiently.</li> | + | <li>We always incubated our ligation overnight in the fridge – maybe incubating at room temperature would work more efficiently. </li> |
| − | <li>Our antibiotic plates had been in the cold room for a long time – had the antibiotic somehow broken down?</li> | + | <li>Our antibiotic plates had been in the cold room for a long time – had the antibiotic somehow broken down? </li> |
</ul> | </ul> | ||
</p> | </p> | ||
<p> | <p> | ||
| − | Nevertheless, the problem was the ligation step of the Interlab procedures. We considered our options, and decided that we would continue with the Interlab Study, but in a different way: we would construct our three devices using IDT gBlocks, assemble them using the Gibson method, transform into E. coli cells and measure their fluorescence. There were a number of reasons why we chose to do this: time was the main issue | + | Nevertheless, the problem was the ligation step of the Interlab procedures. We considered our options, and decided that we would continue with the Interlab Study, but in a different way: we would construct our three devices using IDT gBlocks, assemble them using the Gibson method, transform into <em>E. coli</em> cells and measure their fluorescence. There were a number of reasons why we chose to do this: time was the main issue. The primary purpose of the Interlab Study is to measure the strength of three promoters in different labs, and we figured that we would be able to fulfil this goal with our new method. We became even more determined to complete the Interlab Study, and re-evaluated our goals: |
<ul> | <ul> | ||
| − | <li>After assembling our three devices, we would measure them using the TECAN microplate reader in our lab.</li> | + | <li>After assembling our three devices, we would measure them using the TECAN and BMG FLUOstar Omega microplate reader in our lab. </li> |
| − | <li>After this, we would perform FACS on our devices to measure fluorescence of each individual cell.</li> | + | <li>After this, we would perform FACS on our devices to measure fluorescence of each individual cell. </li> |
| − | <li>We would subsequently use ImageStream to take pictures of our cells.</li> | + | <li>We would subsequently use ImageStream to take pictures of our cells. </li> |
| − | <li>Expressing the Interlab Study constructs cell-free would be another one of our goals: this would allow us to compare fluorescence in cells and cell-free.</li> | + | <li>Expressing the Interlab Study constructs cell-free would be another one of our goals: this would allow us to compare fluorescence in cells and cell-free. </li> |
| − | Full of hope and determination, we ordered the gBlocks that would later be assembled into our three devices. | + | Full of hope and determination, we ordered the gBlocks that would later be assembled into our three devices. |
</p> | </p> | ||
| Line 425: | Line 433: | ||
<div class="ch5"> | <div class="ch5"> | ||
| − | <h2>Chapter 5: Seeing Red</h2> | + | <h2>Chapter 5: Seeing Red</h2> |
<p> | <p> | ||
| − | It took a few weeks for the gBlocks to arrive in our lab. We used this time to prepare for our next attempt at the Interlab study – plan our experiments, find and understand the protocols, especially ones relating to gBlock assembly, which none of us had done before. Once the sequences arrived, a dedicated team of three – Bradley, Jasmine and Georgina – went to the lab to attack the Interlab Study again. gBlocks were prepped and constructed according to the protocols, transformed into DH5α cells, then plated and incubated overnight. Our standard positive and negative controls were used, as well as the NEB positive control from the gBlock assembly kit. | + | It took a few weeks for the gBlocks to arrive in our lab. We used this time to prepare for our next attempt at the Interlab study – plan our experiments, find and understand the protocols, especially ones relating to gBlock assembly, which none of us had done before. Once the sequences arrived, a dedicated team of three – Bradley, Jasmine and Georgina – went to the lab to attack the Interlab Study again. gBlocks were prepped and constructed according to the protocols, transformed into DH5α cells, then plated and incubated overnight. Our standard positive and negative controls were used, as well as the NEB positive control from the gBlock assembly kit. |
</p> | </p> | ||
<p> | <p> | ||
| − | Life is full of surprises, as our team found out the next morning. For the first time, we saw glowing colonies – but they were not glowing green. They were red. We figured out that the plasmid in which our Interlab sequences were in, pSB1C3, which is made from RFP parts, had reformed. | + | Life is full of surprises, as our team found out the next morning. For the first time, we saw glowing colonies – but they were not glowing green. They were red. We figured out that the plasmid in which our Interlab sequences were in, pSB1C3, which is made from RFP parts, had reformed. |
</p> | </p> | ||
| − | + | <center> <img src= "https://static.igem.org/mediawiki/2015/9/94/Exeter_2015_-_ILS_red_colonies.JPG" height=300px> </center> | |
| − | + | ||
<p> | <p> | ||
| − | In the afternoon, two of us went back to the lab, and picked five non-red colonies and one red colony from the plates for each device, as well as one colony from a positive control. We were hoping to see some green glowing after growing them out overnight. However, the colonies we got back were limited in number and did not glow. Looking for an explanation, we decided to miniprep samples from each device, and sequence them to find out exactly what had happened. | + | In the afternoon, two of us went back to the lab, and picked five non-red colonies and one red colony from the plates for each device, as well as one colony from a positive control. We were hoping to see some green glowing after growing them out overnight. However, the colonies we got back were limited in number and did not glow. Looking for an explanation, we decided to miniprep samples from each device, and sequence them to find out exactly what had happened. |
</p> | </p> | ||
| Line 454: | Line 461: | ||
<div class="ch6"> | <div class="ch6"> | ||
| − | <h2>Chapter 6: | + | <h2>Chapter 6: Victory</h2> |
| + | <p> To begin our measurements we thought we would look at whether our ILS constructs displayed any fluorescence at all. To do this we used every machine we could lay our hands on, including FACS, Imagestream, TECAN and the FLUOstar Omega microplate reader kindly lent to us by BMG Labtech. At the beginning of the measurements we also planned several other things we could test our ILS constructs on/in, since our main project was cell free, we thought we should also test the ILS constructs cell free. As an ambitious team, this of course was not the only extra testing we wanted to do with our newly fluorescing constructs, which we had worked so hard to get to this stage. Other plans included growing up in not one, not two but three different ways! In microplate wells, in erlenmeyer flasks and finally in baffled flasks. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | To obtain quantitative fluorescence values for D1, D2 and D3 we used FACS - Fluorescence Activated Cell Sorting. This allowed us to measure the fluorescence of 10,000 individual cells from each device, some of which were glowing more strongly than others, allowing us to see the cell to cell variation for each device. We then used Imagestream to take photos of each cell as we thought it would be useful in complementing and discussing our data. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | To ensure our data was consistent, we made the decision to test the constructs on two different microplate readers in the lab. One of these was the the FLUOstar microplate reader, lent to us by BMG labtech, and the other was a TECAN microplate reader our lab had. The purpose of running the constructs on two different machines was to compare the same samples using different running protocols. As expected, this data was consistent between readers. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | Since our project involved expression of GFP cell free, we thought it would be interesting to see how well other constructs which express GFP would work in a cell free system. Surprisingly, there was a difference between the data collected from expression of the devices in cells, and expression cell free. We thought this could be due to a difference in conditions. This has helped further characterise the parts for the registry. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | |||
| + | We would have liked to test the constructs in different media (LB vs. Minimal media) as well as in shaking flasks, baffled flasks and microplates. We managed to grow up the constructs in microplates and measure the OD600 and fluorescence of these over time, unfortunately we did not have time to run the other conditions. | ||
| + | </p> | ||
</div> | </div> | ||
| Line 464: | Line 490: | ||
<div class="ch7"> | <div class="ch7"> | ||
| − | <h2>Chapter 7: | + | |
| + | <h2>Chapter 7: Conclusion</h2> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | We had many difficulties reaching a point where we could measure the strength of the promoters, it was a very useful experience, especially for technical lab practice, and even though contributing in the interlab study cut into time for our main project, we are happy that we have been able to help contribute further to synthetic biology. | ||
| + | </p> | ||
| + | |||
| + | <center><img src="https://static.igem.org/mediawiki/2015/3/3e/Exeter_2015_-_ILS_lab_team_pic.PNG" height=300px width=400px/></center> | ||
| + | |||
| + | |||
| + | |||
</div> | </div> | ||
| + | |||
<!--End chapter 7--> | <!--End chapter 7--> | ||
| Line 477: | Line 515: | ||
<p> | <p> | ||
| − | + | As has been explained in the previous sections, the ILS devices were measured in a few different ways. Here the data collected is analysed and discussed. | |
</p> | </p> | ||
<h4>Data analysis from microplate readers over 24 hours of growth:</h4> | <h4>Data analysis from microplate readers over 24 hours of growth:</h4> | ||
| + | |||
| + | |||
| + | <div class="containerSlider300" style="float:right"> | ||
| + | <div class="slider300"> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/9/9d/Exeter_FLUOstar_OD600_fluorescence_blank_corrected.png"><img src="https://static.igem.org/mediawiki/2015/9/9d/Exeter_FLUOstar_OD600_fluorescence_blank_corrected.png" title="Figure 4: Graph of time vs. blank-corrected fluorescence of ILS devices as read using BMG FLUOstar Omega plate reader."></a></div> | ||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/9/9d/Exeter_FLUOstar_OD600_fluorescence_blank_corrected.png"><img src="https://static.igem.org/mediawiki/2015/9/9d/Exeter_FLUOstar_OD600_fluorescenceactual_blank_corrected.png" title="Figure 4: Graph of time vs. blank-corrected OD<sub>600</sub> ILS devices as read using BMG FLUOstar Omega plate reader."></a></div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
<p> | <p> | ||
| + | Fluorescence readings for the ILS devices expressed in cells were gained using the protocol found <a href="">here</a>. Briefly, overnight cultures from glycerol stocks of the three devices, R0040, I20270, and untransformed DH5α <em>E.coli</em> cells (cell growth control) were diluted to an OD<sub>600</sub> of 0.1 and loaded into a 96-well microplate using an algorithm to ensure minimal cross-talk between samples. Two microplates were set up; one was read by a BMG FLUOstar Omega plate reader, and the other by a Tecan plate reader. The plates were incubated with shaking at 37C and read every 60 mins over a 24 hour period, however due to an equipment failure data was lost between hours 10 and 22 for the BMG reader, and between hours 7 and 21 for the Tecan reader. At each read, GFP fluorescence and OD<sub>600</sub> was measured and recorded. | ||
| + | </p> | ||
| + | |||
| + | <div class="containerSlider300" style="float:left"> | ||
| + | <div class="slider300"> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/7/7f/Exeter_tecan_OD600_fluorescence_blank_corrected.png"><img src="https://static.igem.org/mediawiki/2015/7/7f/Exeter_tecan_OD600_fluorescence_blank_corrected.png" title="Figure 5: Graph of time vs. blank-corrected OD<sub>600</sub> and fluorescence of ILS devices as read using Tecan plate reader."></a></div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | The data collected from each reader was blank-corrected before being plotted as time vs. fluorescence intensity (AU), and time vs. OD<sub>600</sub> (i.e. cell growth). Values for GFP fluorescence intensity at an OD<sub>600</sub> of 0.5 were deduced from the graphs and submitted to allow for comparison of data between other teams/labs. These graphs, along with the values at OD<sub>600</sub> of 0.5, are shown in figures 1 and 2. As can be seen, all three devices show lower GFP production than the positive control (I20270), with D2 showing the strongest fluorescence and D1 showing the weakest, excepting the R0040 negative GFP control. </br> | ||
| + | As can also be seen from both graphs, GFP fluorescence intensity follows roughly the same trend as OD<sub>600</sub> (i.e. cell growth), which generally increases over time before plateauing. | ||
</p> | </p> | ||
| − | <h4>Data analysis from FACS ( | + | </br> |
| + | </br> | ||
| + | </br> | ||
| + | |||
| + | <h4>Data analysis from FACS (fluorescence associated cell sorting) and ImageStream:</h4> | ||
| + | |||
| + | <div class="containerSlider500" style="float:right;"> | ||
| + | <div class="slider500"> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/9/9d/Exeter_I20270_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/9/9d/Exeter_I20270_FACS_histogram.png" title="Figure 3: FACS histogram for I20270 cells (GFP positive control)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/0/01/Exeter_R0040_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/0/01/Exeter_R0040_FACS_histogram.png" title="Figure 3: FACS histogram for R0040 cells (GFP negative control)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/1/11/Exeter_D1a_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/1/11/Exeter_D1a_FACS_histogram.png" title="Figure 3: FACS histogram for Device 1a cells (biological replicate 1)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/1/1a/Exeter_D1b_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/1/1a/Exeter_D1b_FACS_histogram.png" title="Figure 3: FACS histogram for Device 1b cells (biological replicate 2)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/2/27/Exeter_D1c_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/2/27/Exeter_D1c_FACS_histogram.png" title="Figure 3: FACS histogram for Device 1c cells (biological replicate 3)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/c/c9/Exeter_D2a_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/c/c9/Exeter_D2a_FACS_histogram.png" title="Figure 3: FACS histogram for Device 2a cells (biological replicate 1)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/b/b7/Exeter_D2b_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/b/b7/Exeter_D2b_FACS_histogram.png" title="Figure 3: FACS histogram for Device 2b cells (biological replicate 2)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/c/c2/Exeter_D2c_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/c/c2/Exeter_D2c_FACS_histogram.png" title="Figure 3: FACS histogram for Device 2c cells (biological replicate 3)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/c/c2/Exeter_D3a_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/c/c2/Exeter_D3a_FACS_histogram.png" title="Figure 3: FACS histogram for Device 3a cells (biological replicate 1)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/1/10/Exeter_D3b_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/1/10/Exeter_D3b_FACS_histogram.png" title="Figure 3: FACS histogram for Device 3b cells (biological replicate 2)."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/f/fe/Exeter_D3c_FACS_histogram.png"><img src="https://static.igem.org/mediawiki/2015/f/fe/Exeter_D3c_FACS_histogram.png" title="Figure 3: FACS histogram for Device 3c cells (biological replicate 3)."></a></div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <p> | ||
| + | To complement the data collected from the plate readers, overnight cultures of the devices (D1, D2, and D3), I20270, R0040, and DH5α cells were measured using FACS and single cells viewed using ImageStream. In order to generate data, fluorescence was measured for 10,000 individual cells and reported as a histogram of fluorescence intensity vs. cell count, and a scattergram of forward light scatter vs. fluorescence intensity. Shown in figure 3 are the FACS histograms for all constructs measured. In order to analyse the activity of the three devices, a region (shown on the histograms as green) was defined covering only ~5% of R0040 cells. This region was then applied to all other FACS histograms for effective comparison between samples, and cells within this region are defined as having fluorescence above baseline. As can be seen in image 4, 99.7% of cells are within this region. In comparison, Device 1 shows an average of 16.8% of cells within the region of fluorescence, as measured over three biological replicates.</br> | ||
| + | |||
| + | <div class="containerSlider300" style="float:left"> | ||
| + | <div class="slider300"> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/f/fa/Exeter_I20270_Imagestream.png"><img src="https://static.igem.org/mediawiki/2015/f/fa/Exeter_I20270_Imagestream.png" title="Figure 4: Image of I20270 cells (GFP positive control) from Imagestream."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/c/c8/Exeter_R0040_Imagestream.png"><img src="https://static.igem.org/mediawiki/2015/c/c8/Exeter_R0040_Imagestream.png" title="Figure 4: Image of R0040 cells (GFP negative control) from Imagestream."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/6/66/Exeter_D1a_Imagestream.png"><img src="https://static.igem.org/mediawiki/2015/6/66/Exeter_D1a_Imagestream.png" title="Figure 4: Image of device 1 cells from Imagestream."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/7/7e/Exeter_D2a_Imagestream.png"><img src="https://static.igem.org/mediawiki/2015/7/7e/Exeter_D2a_Imagestream.png" title="Figure 4: Image of device 2 cells from Imagestream."></a></div> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/a/a6/Exeter_D3a_Imagestream.png"><img src="https://static.igem.org/mediawiki/2015/a/a6/Exeter_D3a_Imagestream.png" title="Figure 4: Image of device 2 cells from Imagestream."></a></div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | Unlike the other cells measured, device 2 doesn't show a strong normal distribution with a single population of cells, but instead shows two peaks representing two main populations. The first and smaller of the populations does not show significant fluorescence (i.e. it is outside of the defined region), however the second and larger of the populations does show significant fluorescence, and overall there is an average of 87.1% of cells showing fluorescence as measured across three biological replicates of device 2.</br> | ||
| + | |||
| + | |||
| + | |||
| + | Finally, Device 3 shows an average of 32.2% of cells with significant fluorescence as compared to the negative GFP control (R0040). This places device 3 between device 2 (highest) and device 1 (lowest) in terms of GFP expression.</br> | ||
| + | Shown in figure 4 are images taken by ImageStream of the three devices, along with I20270 and R0040. The images were all viewed in the same way (the same pixel view, PMT, etc.) to allow for comparison between samples. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <div class="containerSlider300" style="float:right"> | ||
| + | <div class="slider300"> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/7/75/Exeter_FLUOstar_cellfree_negative_corrected.png"><img src="https://static.igem.org/mediawiki/2015/7/75/Exeter_FLUOstar_cellfree_negative_corrected.png" title="Figure 5: Chart of negative-corrected fluorescence intensity for all ILS constructs expressed in a Promega S30 cell free kit after 60 mins incubation."></a></div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <h4>Cell-free expression in an S30 cell-free kit:</h4> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <p> | ||
| + | In order to help characterise the Promega S30 cell-free kit for use in our project, the three devices plus the positive and negative controls were expressed cell-free. The exact protocol followed can be found <a href="">here</a>, but briefly ~1μg of plasmid DNA of each construct was added to a promega S30 kit and incubated at 37C for 1 hour, after which the reaction was stopped by placing on ice and read in by the BMG plate reader. Shown in figure 5 is a chart of fluorescence of each construct corrected so that R0040 has a fluorescence of 0. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h4>Discussion of results:</h4> | ||
| + | |||
| + | <p> | ||
| + | As can be seen, the strength of the promoters measured are as follows from lowest to highest; J23101, J23117, J23106. This is not what would be expected according to data in the iGEM registry on each of these promoters, which have already had strength measured using RFP as the reporter. According to the data in the registry, the strength of promoters from lowest to highest should be; J23117, J23106, J23101. The strengths measured for J23117 and J23106 (devices 2 and 3) are as would be expected, however J23101 is not. This would suggest that during the process of device construction, or the process of growing up and expressing the promoter within cells during measurement, the J23101 promoter became damaged or otherwise affected in a negative way. Sequencing data shows that the promoter was not mutated in any way, therefore suggesting that the cells and conditions in which device 1 was grown and expressed were unfavourable for the J23101 promoter.</br> | ||
| + | </br> | ||
| + | The data which was collected by our team was mainly consistent, i.e. all forms of data collected suggests that J23106 (device 2) is the strongest promoter, followed by J23117 (device 3), and finally J23101 (device 1), with the GFP positive control (I20270) showing higher GFP production than any of the devices. However, the data collected from cell free expression of the constructs goes against the other data collected. Cell-free, the strength of the promoters from strongest to weakest is J23101, J23117, J23106, with J23101 showing higher activity than the positive control (I20270). The data collected cell free matches that which would be expected from data already in the iGEM registry. This matches the hypothesis stated above that the reason J23106 is weaker than expected is due to the growth conditions of the cells expressing device 2, or the strain of cells in which it is being expressed, as neither of these factors occur in cell-free expression. This suggests that the BioBrick J23106 may not be very standard between labs/growth or expression conditions. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <!--End epilogue--> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--Start protocols--> | ||
| + | |||
| + | |||
| + | <div class="proto" style="min-height:1000px;"> | ||
| + | |||
| + | <h2>Supplementary Information</h2> | ||
| + | |||
| + | <!-- | ||
<div class="containerSlider500" style="float:left;"> | <div class="containerSlider500" style="float:left;"> | ||
| Line 563: | Line 738: | ||
</div> | </div> | ||
| + | --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | <h4>Supplementary download 1 - Sequencing data:</h4> | ||
| + | <div class="containerSlider300" style="float:left"> | ||
| + | <div class="sliderstatic300"> | ||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/a/a9/ILS_sequence_files.zip"><img src="https://static.igem.org/mediawiki/2015/5/5b/Exeter_ZIPImage_ILS_Sequencing.png" title="Click the image to download all ILS sequencing data."></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | <h4>Supplementary download 2 - data files:</h4> | ||
| + | <p> | ||
| − | < | + | <a href="https://static.igem.org/mediawiki/2015/5/5f/Exeter_ILS_raw.zip">Click here to download</a> |
| + | </p> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| − | |||
| − | < | + | <!--End protocols--> |
| + | </div> | ||
| + | <!-- This is where my buttons are --> | ||
| − | < | + | <nav> |
| + | <ul class="pager"> | ||
| + | <li class="previous"><a href="https://2015.igem.org/Team:Exeter/Safety"><span aria-hidden="true">←</span>Safety</a></li> | ||
| + | <li class="next"><a href="https://2015.igem.org/Team:Exeter/Modeling">Modelling<span aria-hidden="true">→</span></a></li> | ||
| + | </ul> | ||
| + | </nav> | ||
| + | <!-- Buttons stop --> | ||
| − | |||
</html> | </html> | ||
{{Exeterfooter}} | {{Exeterfooter}} | ||
Latest revision as of 03:43, 19 September 2015
Interlab Study
Prologue: The Devices
The aim of the interlab study was to measure the strength of three constitutive Anderson promoters, using GFP as a measure of strength, in order to generate data which can then be compared between different iGEM labs. Here we tell our story of the interlab study, including the promoters which were tested, and the ways in which we measured their strength. Follow the chapters in order using the buttons above to be guided through this story. We begin in this section by stating the promoters which were measured, and the devices these were assembled into in order to measure them, along with the controls used.
Devices 1, 2 and 3:
Three devices were constructed in order to test the promoters; D1, D2 and D3. Each device contains a promoter, followed by the GFP measurement composite part I13504 (figure 1). This part contains the GFP biobrick BBa_E0040, which is preceded by a strong RBS (B0034) and succeeded by a double terminator (B0010 and B0012). By using this same part in all of the devices constructed, it is ensured that the only variation between devices is the promoter, and the use of a strong RBS ensures that the expression of GFP should be limited by the strength of the promoter, allowing measurement of promoter strength.
The promoters used in devices 1, 2 and 3 were J23101 (D1), J23106 (D2), and J23117 (D3) respectively (figure 1), and all devices were built into and expressed in a pSB1C3 plasmid backbone. The pSB1C3 plasmid confers chloramphenicol resistance, which was used to select for cells containing the device. Once built, all of the devices were sequenced verified (figure 2, also available for download here).
The Controls:
In the measurement of the above devices, three controls were used; a GFP positive control (I20270), a GFP negative control (R0040), and a cell growth control (untransformed cells). I20270 contains the same RBS, GFP gene, and double terminator, but contains the J23151 promoter. J23151 is also a constitutive promoter, allowing for a direct comparison to the promoters in the devices built, as well as acting as a positive GFP control. R0040 contains a Tet repressible promoter, meaning that it will not fluoresce and can act as a GFP negative control during measurement. Figure 3 shows schematics of these parts.
Chapter 1: New Beginnings
Once upon a time, there was an iGEM team from the University of Exeter. Determined to contribute to the field of synthetic biology, we decided to participate in the Second International Interlab Measurement Study. While determining fluorescence levels for the three Interlab devices, we hoped to gain some experience in the lab before starting our iGEM project, Ribonostics. We aimed to build and measure the fluorescence of three following BioBrick devices into pSB1C3 plasmid backbone:
- J23101 + I13504 (D1)
- J23106 + I13504 (D2)
- J23117 + I13504 (D3)
In the first week of our iGEM induction, we started out by re-hydrating the required DNA from the kit, after which we followed the standard protocols for BioBrick assembly. The first step of this process was transforming the BioBrick DNA into competent DH5α cells (from New England Biolabs, NEB), before growing them up on agar plates containing the correct antibiotic (ampicillin for I13504, chloramphenicol in the promoters). We then prepared overnight cultures. After transforming, we performed our first miniprep to obtain the DNA needed for further experiments, and checked the DNA concentration using Qubit. We then used values obtained from this to calculate the volumes of all of components of the digestion step. After the digestion, we ran a part of each sample on a gel to see if the insert and vector were the correct size. The ligation was the last step; after this, we plated our samples and incubated them overnight. We hoped that the next morning, there would be glowing green colonies on our plates. Protocols of these experiments can be found here : transformation, miniprep, Qubit, digestion, ligation.
The next day, we came into the lab, hoping to see colonies so green they would be blinding. After viewing the plates under UV light to visualise the green fluorescent protein (GFP) in our constructs, we understood that the statement ‘biology sometimes doesn’t work’ is entirely accurate. There were no glowing colonies on our plates. However, this moment of truth inspired us to diagnose what was wrong with our devices, and give the Interlab Study another try in the following week.
Our lab induction and Interlab Study were a good chance to find out where the equipment and resources in the lab were, as well as to meet some of the researchers working there – many of them helped at various stages of our project. Those amongst us with little or no lab experience learned basic techniques, such as pouring agar and making LB broth, but also made an effort to understand the biological concepts behind the Interlab Study. One of our physicists, Todd, said that in the first week, he learned the skills necessary for any budding biologist – pipetting, making, and pouring agar. ‘I thought it was very useful. It was great.’ – a direct quote from Todd. Equipped with these skills, we set out to complete the Interlab Study accurately, efficiently, and most importantly – successfully.
Chapter 2: Under Pressure
After a weekend of thought, we came into the lab, determined to get glowing colonies by the end of the week. The answer to why our interlab study failed the previous week was simple: the promoters we were using were not BioBrick assembly compatible – they did not join, because they could not join.
We looked at our second attempt at the interlab study with optimism – we had some practice in the lab, and we understood more clearly what it was we were actually trying to do. This time, we made and used TOP10 competent cells. Again, we followed the same flowchart of experiments, using the correct BioBricks (J23101, J23106, J23117).
This time, only three or four of us worked in the lab at any one time. During our first week in the lab, we learned that having more people in the lab does not necessarily mean we are more productive. During the second try at the Interlab Study, we also made sure we had all the necessary controls – our competent TOP10 cells alone, the empty pSB1C3 backbone, a working GFP plasmid and a negative control of the TetR plasmid. After a few setbacks (such as forgetting to put LB broth during the incubation stage of a bacterial transformation), we arrived at yet another overnight incubation, and again, we asked ourselves the burning question – would we see glowing colonies?
Chapter 3: We Fight On
By now, you may have guessed that our team is not the luckiest when it comes to glowing. If so, you guessed right – we did not have any glowing colonies.
We were baffled, puzzled, surprised – but not broken. Once again, we marched into the lab, determined to see fluorescent colonies. We double (and triple) checked that we were using the correct BioBricks, and enzymes during the digestion step. We also thought carefully about what we could change in order to get our Interlab Study to work. During our third attempt, we followed a protocol for a traditional digestion instead of the digestion protocol we had used before. We also altered the transformation protocol slightly. After running our digest on the gel, we performed a gel extraction – this way we would be sure that we were using vector and insert fragments of the correct sizes. Amongst the things we added and changed were also: control plates of just our competent cells, and using 1 µl of fresh T4 ligase instead of 0.5 µl. Again, we followed the Interlab flowchart of procedures. Armed with our transformed Interlab samples and plenty of controls, we saw no reason for our colonies not to glow the next morning.
Chapter 4: New Direction
On the rainy Friday morning, we climbed the stairs to the fourth floor, where our lab was located. We had great – but realistic – expectations for our colonies. Once we saw them, we knew the likely result of our third attempt at the Interlab Study. The UV light confirmed our suspicions – we did not have any green glowing colonies. We had a discussion about what could be going wrong with the Interlab process, and came up with three hypotheses:
- During our ligation, we used a 1:1 ratio of insert to vector – perhaps using a 3:1 or 1:3 ratio would be better.
- We always incubated our ligation overnight in the fridge – maybe incubating at room temperature would work more efficiently.
- Our antibiotic plates had been in the cold room for a long time – had the antibiotic somehow broken down?
Nevertheless, the problem was the ligation step of the Interlab procedures. We considered our options, and decided that we would continue with the Interlab Study, but in a different way: we would construct our three devices using IDT gBlocks, assemble them using the Gibson method, transform into E. coli cells and measure their fluorescence. There were a number of reasons why we chose to do this: time was the main issue. The primary purpose of the Interlab Study is to measure the strength of three promoters in different labs, and we figured that we would be able to fulfil this goal with our new method. We became even more determined to complete the Interlab Study, and re-evaluated our goals:
- After assembling our three devices, we would measure them using the TECAN and BMG FLUOstar Omega microplate reader in our lab.
- After this, we would perform FACS on our devices to measure fluorescence of each individual cell.
- We would subsequently use ImageStream to take pictures of our cells.
- Expressing the Interlab Study constructs cell-free would be another one of our goals: this would allow us to compare fluorescence in cells and cell-free. Full of hope and determination, we ordered the gBlocks that would later be assembled into our three devices.
Chapter 5: Seeing Red
It took a few weeks for the gBlocks to arrive in our lab. We used this time to prepare for our next attempt at the Interlab study – plan our experiments, find and understand the protocols, especially ones relating to gBlock assembly, which none of us had done before. Once the sequences arrived, a dedicated team of three – Bradley, Jasmine and Georgina – went to the lab to attack the Interlab Study again. gBlocks were prepped and constructed according to the protocols, transformed into DH5α cells, then plated and incubated overnight. Our standard positive and negative controls were used, as well as the NEB positive control from the gBlock assembly kit.
Life is full of surprises, as our team found out the next morning. For the first time, we saw glowing colonies – but they were not glowing green. They were red. We figured out that the plasmid in which our Interlab sequences were in, pSB1C3, which is made from RFP parts, had reformed.

In the afternoon, two of us went back to the lab, and picked five non-red colonies and one red colony from the plates for each device, as well as one colony from a positive control. We were hoping to see some green glowing after growing them out overnight. However, the colonies we got back were limited in number and did not glow. Looking for an explanation, we decided to miniprep samples from each device, and sequence them to find out exactly what had happened.
So once again the fluorescence promised to us by our interlab constructs had eluded us again, since we knew the wait for our sequencing results could take a while, we decided to move on and start with the gBlocks from scratch. However half way through this process we received the results back from our first gBlock sequences and it had worked! This left us with more questions, if the gBlocks had worked then why were they not glowing? So we proceeded to use a different machine to measure fluorescence, picking TECAN which showed clearly that they did indeed fluoresce just not as brightly as we had expected. With this in mind we started to plan all the things we could measure.
Chapter 6: Victory
To begin our measurements we thought we would look at whether our ILS constructs displayed any fluorescence at all. To do this we used every machine we could lay our hands on, including FACS, Imagestream, TECAN and the FLUOstar Omega microplate reader kindly lent to us by BMG Labtech. At the beginning of the measurements we also planned several other things we could test our ILS constructs on/in, since our main project was cell free, we thought we should also test the ILS constructs cell free. As an ambitious team, this of course was not the only extra testing we wanted to do with our newly fluorescing constructs, which we had worked so hard to get to this stage. Other plans included growing up in not one, not two but three different ways! In microplate wells, in erlenmeyer flasks and finally in baffled flasks.
To obtain quantitative fluorescence values for D1, D2 and D3 we used FACS - Fluorescence Activated Cell Sorting. This allowed us to measure the fluorescence of 10,000 individual cells from each device, some of which were glowing more strongly than others, allowing us to see the cell to cell variation for each device. We then used Imagestream to take photos of each cell as we thought it would be useful in complementing and discussing our data.
To ensure our data was consistent, we made the decision to test the constructs on two different microplate readers in the lab. One of these was the the FLUOstar microplate reader, lent to us by BMG labtech, and the other was a TECAN microplate reader our lab had. The purpose of running the constructs on two different machines was to compare the same samples using different running protocols. As expected, this data was consistent between readers.
Since our project involved expression of GFP cell free, we thought it would be interesting to see how well other constructs which express GFP would work in a cell free system. Surprisingly, there was a difference between the data collected from expression of the devices in cells, and expression cell free. We thought this could be due to a difference in conditions. This has helped further characterise the parts for the registry.
We would have liked to test the constructs in different media (LB vs. Minimal media) as well as in shaking flasks, baffled flasks and microplates. We managed to grow up the constructs in microplates and measure the OD600 and fluorescence of these over time, unfortunately we did not have time to run the other conditions.
Chapter 7: Conclusion
We had many difficulties reaching a point where we could measure the strength of the promoters, it was a very useful experience, especially for technical lab practice, and even though contributing in the interlab study cut into time for our main project, we are happy that we have been able to help contribute further to synthetic biology.

Epilogue: Results
As has been explained in the previous sections, the ILS devices were measured in a few different ways. Here the data collected is analysed and discussed.
Data analysis from microplate readers over 24 hours of growth:
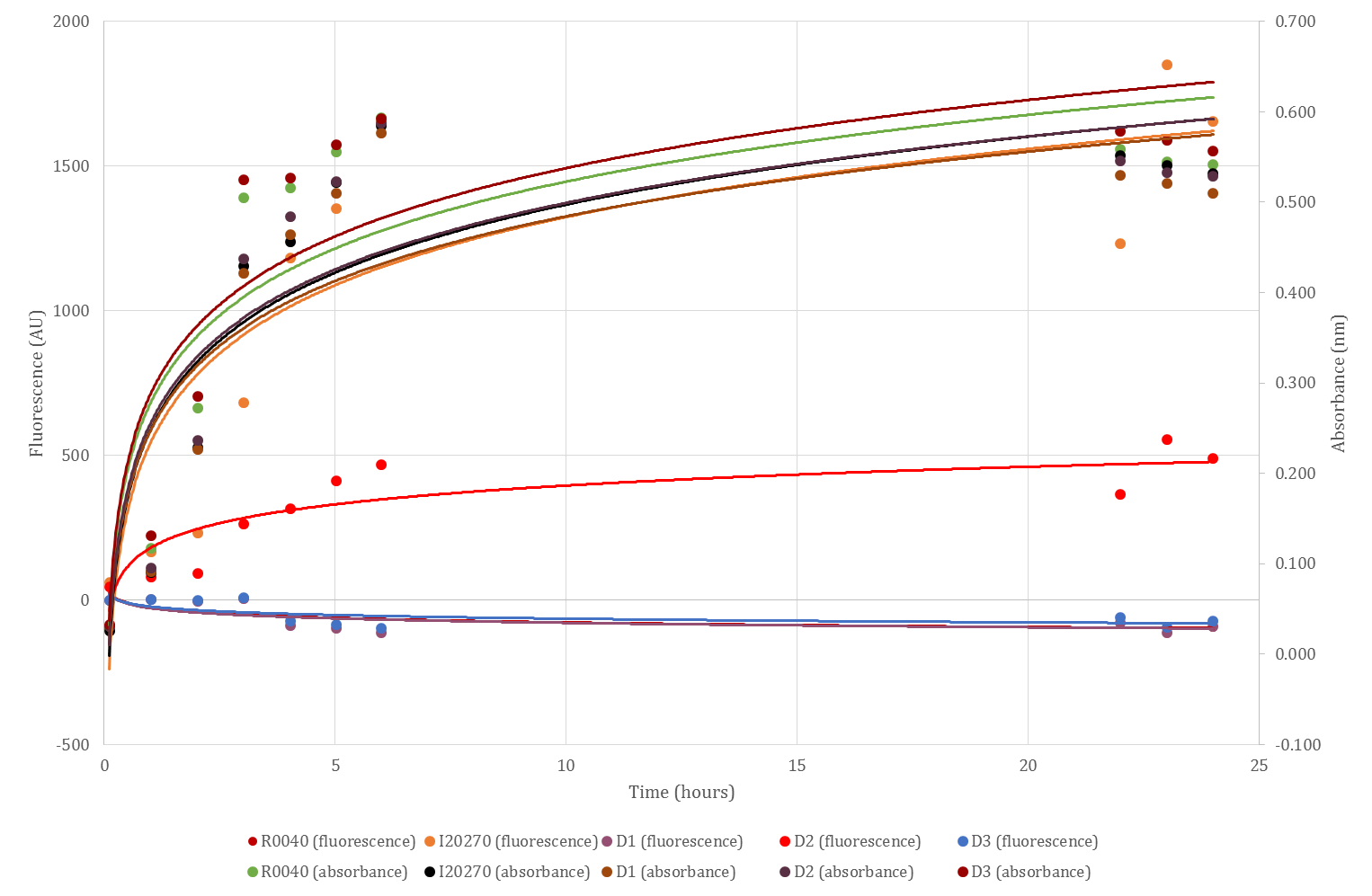
Fluorescence readings for the ILS devices expressed in cells were gained using the protocol found here. Briefly, overnight cultures from glycerol stocks of the three devices, R0040, I20270, and untransformed DH5α E.coli cells (cell growth control) were diluted to an OD600 of 0.1 and loaded into a 96-well microplate using an algorithm to ensure minimal cross-talk between samples. Two microplates were set up; one was read by a BMG FLUOstar Omega plate reader, and the other by a Tecan plate reader. The plates were incubated with shaking at 37C and read every 60 mins over a 24 hour period, however due to an equipment failure data was lost between hours 10 and 22 for the BMG reader, and between hours 7 and 21 for the Tecan reader. At each read, GFP fluorescence and OD600 was measured and recorded.
The data collected from each reader was blank-corrected before being plotted as time vs. fluorescence intensity (AU), and time vs. OD600 (i.e. cell growth). Values for GFP fluorescence intensity at an OD600 of 0.5 were deduced from the graphs and submitted to allow for comparison of data between other teams/labs. These graphs, along with the values at OD600 of 0.5, are shown in figures 1 and 2. As can be seen, all three devices show lower GFP production than the positive control (I20270), with D2 showing the strongest fluorescence and D1 showing the weakest, excepting the R0040 negative GFP control. As can also be seen from both graphs, GFP fluorescence intensity follows roughly the same trend as OD600 (i.e. cell growth), which generally increases over time before plateauing.
Data analysis from FACS (fluorescence associated cell sorting) and ImageStream:
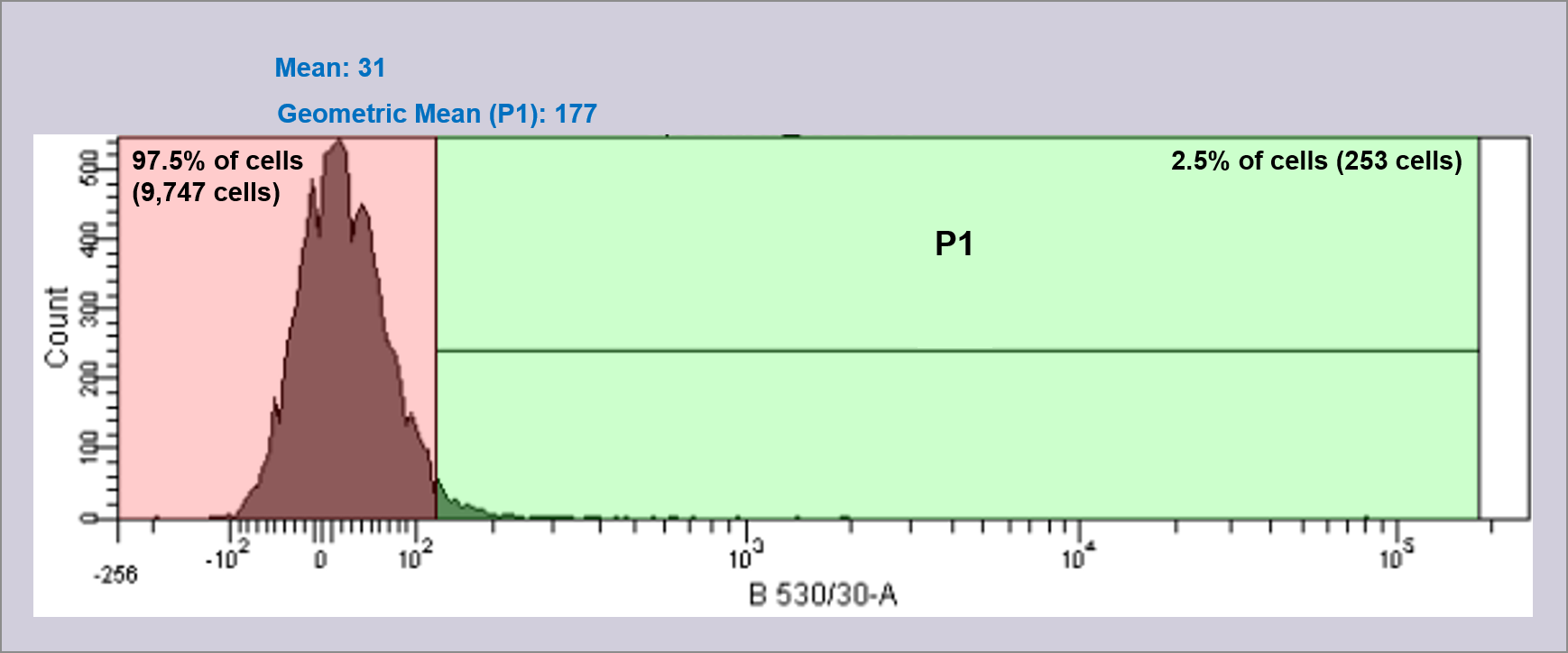
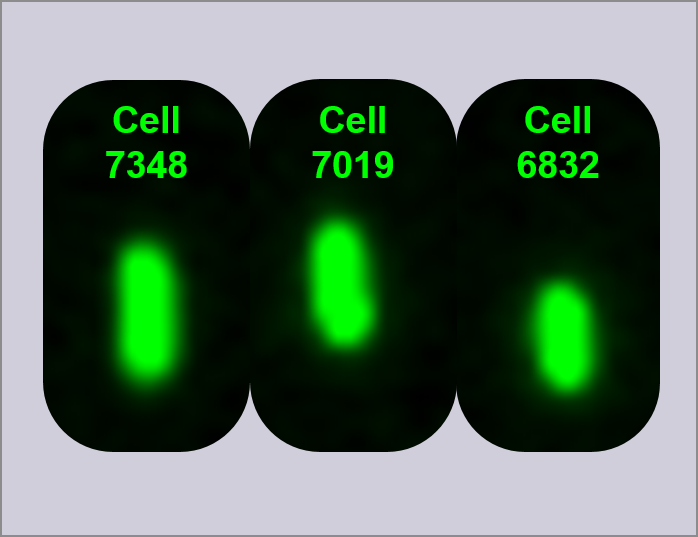
To complement the data collected from the plate readers, overnight cultures of the devices (D1, D2, and D3), I20270, R0040, and DH5α cells were measured using FACS and single cells viewed using ImageStream. In order to generate data, fluorescence was measured for 10,000 individual cells and reported as a histogram of fluorescence intensity vs. cell count, and a scattergram of forward light scatter vs. fluorescence intensity. Shown in figure 3 are the FACS histograms for all constructs measured. In order to analyse the activity of the three devices, a region (shown on the histograms as green) was defined covering only ~5% of R0040 cells. This region was then applied to all other FACS histograms for effective comparison between samples, and cells within this region are defined as having fluorescence above baseline. As can be seen in image 4, 99.7% of cells are within this region. In comparison, Device 1 shows an average of 16.8% of cells within the region of fluorescence, as measured over three biological replicates.
Cell-free expression in an S30 cell-free kit:
In order to help characterise the Promega S30 cell-free kit for use in our project, the three devices plus the positive and negative controls were expressed cell-free. The exact protocol followed can be found here, but briefly ~1μg of plasmid DNA of each construct was added to a promega S30 kit and incubated at 37C for 1 hour, after which the reaction was stopped by placing on ice and read in by the BMG plate reader. Shown in figure 5 is a chart of fluorescence of each construct corrected so that R0040 has a fluorescence of 0.
Discussion of results:
As can be seen, the strength of the promoters measured are as follows from lowest to highest; J23101, J23117, J23106. This is not what would be expected according to data in the iGEM registry on each of these promoters, which have already had strength measured using RFP as the reporter. According to the data in the registry, the strength of promoters from lowest to highest should be; J23117, J23106, J23101. The strengths measured for J23117 and J23106 (devices 2 and 3) are as would be expected, however J23101 is not. This would suggest that during the process of device construction, or the process of growing up and expressing the promoter within cells during measurement, the J23101 promoter became damaged or otherwise affected in a negative way. Sequencing data shows that the promoter was not mutated in any way, therefore suggesting that the cells and conditions in which device 1 was grown and expressed were unfavourable for the J23101 promoter. The data which was collected by our team was mainly consistent, i.e. all forms of data collected suggests that J23106 (device 2) is the strongest promoter, followed by J23117 (device 3), and finally J23101 (device 1), with the GFP positive control (I20270) showing higher GFP production than any of the devices. However, the data collected from cell free expression of the constructs goes against the other data collected. Cell-free, the strength of the promoters from strongest to weakest is J23101, J23117, J23106, with J23101 showing higher activity than the positive control (I20270). The data collected cell free matches that which would be expected from data already in the iGEM registry. This matches the hypothesis stated above that the reason J23106 is weaker than expected is due to the growth conditions of the cells expressing device 2, or the strain of cells in which it is being expressed, as neither of these factors occur in cell-free expression. This suggests that the BioBrick J23106 may not be very standard between labs/growth or expression conditions.