Difference between revisions of "Team:Austin UTexas/Interlab Study"
Kvasudevan (Talk | contribs) |
|||
| (12 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Austin_UTexas}} | {{Austin_UTexas}} | ||
| + | =Interlab Measurement Study= | ||
| − | + | The Interlab Study is an initiative started in 2014 by the iGEM foundation to collect standard measurement data from around the world. This year, as a part of the measurement track, we completed the Interlab study according to the directions provided on the [[Tracks/Measurement/Interlab_study|iGEM Interlab Study webpage]]. The purpose of this year's study was to create three different GFP containing devices through BioBrick assembly and measure the fluorescence across biological replicates. Below, we have detailed our procedures, implications of our results, and conclusions we have drawn that relate back to our main focus of evolutionary stability in synthetic biology. In synthetic biology, it is important that part characterization is consistent between different labs to create well-defined standard parts for the Registry. We are excited to contribute to this study! | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | The Interlab Study is an initiative started in 2014 by the iGEM foundation to collect standard measurement data from around the world. This year, as a part of the measurement track, we completed the Interlab study according to the directions provided on the | + | |
| − | + | ||
| + | ==Protocol== | ||
| + | 1. Transform the genetic parts from the BioBrick 2015 kit into TOP10 ''E. coli'' cells. BioBrick assembly was used to engineer the InterLab study devices. Refer to the BioBrick website for more information on assembly procedure: http://parts.igem.org/Assembly:Standard_assembly. | ||
| − | + | 2. Streak out an agar plate (LB/CAM) with the ''E. coli'' containing the device and any controls | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | 2. Streak out an agar plate (LB/CAM) with the E. coli containing the device and any controls | + | |
<br> | <br> | ||
| Line 35: | Line 25: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | + | <b>Plate Reader Measurement Protocol:</b> | |
<hr> | <hr> | ||
| − | + | <p> | |
- Obtain initial OD600 measurement of O/N cultures | - Obtain initial OD600 measurement of O/N cultures | ||
</p> | </p> | ||
| Line 61: | Line 51: | ||
</p><br> | </p><br> | ||
<br> | <br> | ||
| − | + | <b>Flow Cytometry Protocol:</b> | |
<hr> | <hr> | ||
| − | |||
<p>- Wash culture in PBS to remove LB</p><p> | <p>- Wash culture in PBS to remove LB</p><p> | ||
- Load 150µl of sample into plate</p><p> | - Load 150µl of sample into plate</p><p> | ||
| Line 71: | Line 60: | ||
<br> | <br> | ||
| − | + | <b>Extra Credit:</b> | |
<hr> | <hr> | ||
| − | |||
<p>Three technical replicates of the experiment were recorded where the measurements were repeated. The experimental samples were measured at least three times with both the plate reader and flow cytometer. Furthermore, we conducted three days of measurements to observe any possible genetic breaking of the devices to confirm our hypothesis that the strongest promoter will confer the most metabolic load on the organism, which can cause loss-of-function mutations within the plasmid. | <p>Three technical replicates of the experiment were recorded where the measurements were repeated. The experimental samples were measured at least three times with both the plate reader and flow cytometer. Furthermore, we conducted three days of measurements to observe any possible genetic breaking of the devices to confirm our hypothesis that the strongest promoter will confer the most metabolic load on the organism, which can cause loss-of-function mutations within the plasmid. | ||
<br> | <br> | ||
<b>Protocol for further measurement of biological triplicates and technical replicates over the course of three days:</b> | <b>Protocol for further measurement of biological triplicates and technical replicates over the course of three days:</b> | ||
| − | + | </p> | |
<br> | <br> | ||
<b>Day 0:</b> | <b>Day 0:</b> | ||
| Line 121: | Line 109: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | + | ==Devices Measured== | |
| − | + | ||
<b>1.</b> The first device was cloned in lab and contains the following BioBrick parts: | <b>1.</b> The first device was cloned in lab and contains the following BioBrick parts: | ||
* A promoter, BBa_J23101, a downstream gene (BBa_I13504), and a backbone (pSB1C3). | * A promoter, BBa_J23101, a downstream gene (BBa_I13504), and a backbone (pSB1C3). | ||
| Line 135: | Line 123: | ||
* The downstream gene is also a composite of a number of BioBrick parts including an RBS - B0034, a GFP coding sequence - E0040, and a transcriptional terminator - B0015. | * The downstream gene is also a composite of a number of BioBrick parts including an RBS - B0034, a GFP coding sequence - E0040, and a transcriptional terminator - B0015. | ||
| + | ==Data and Discussion== | ||
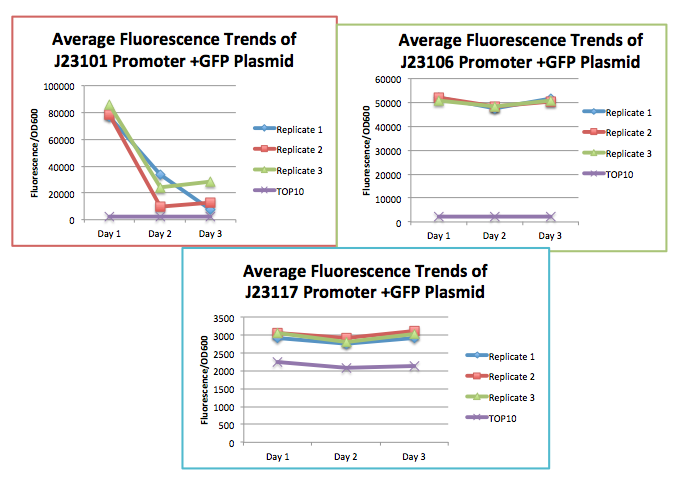
| + | [[Image:2015_Austin_UTexas_Interlab_3GRAPHS.png|500px|thumb|right|<b>Fig. 1:</b> Charts tracking the relative fluorescence of three replicates (1, 2, 3) each for 3 devices: GFP+BBa_J23101, GFP+BBa_J23106, and GFP+BBa_J23117]] | ||
| + | The 2015 Interlab study provided a substantive way to continue our study of evolutionary stability in synthetic organisms. The framework of the study allowed us to understand the differences in metabolic load conferred by varying promoter strengths. All three devices created for this study were composed of the same downstream gene (BBa_I13504), and a backbone (pSB1C3). The order of promoter strength was as follows: J213101, J23106, J23117. The promoters used for the construction of the three devices are constitutive promoters of varying strengths, and should hypothetically generate varying metabolic load on the organism. All devices have the same coding sequence, RBS, transcriptional terminator, backbone, and thus plasmid copy number. We used the Interlab study as means to further our research to continue to measure this fitness cost imposed by identifying any point mutations, small insertions and deletions, large deletions, repeated operator sequences within the promoter, and insertion sequence (IS) element insertions in the sequencing data. It could help the effort to standardize promoter measurements between promoters in the same family, and help strengthen any evolutionary stability arguments made in the Breaking is Bad project. | ||
| − | + | The results of the lab generally followed our hypothesis. As seen from <b>(Fig. 1 and 2)</b>, the J23101 promoter containing plasmid, device 1, with the strongest constitutive expression showed a significant loss in fluorescence after day 1, while the J23106 and J23117 promoter containing plasmids, device 2 and 3, respectively, were able to maintain stable levels of fluorescence over the course of three days. However, we expect these plasmids to eventually break. Although we did not sequence the broken cultures, visual assessment of the plates and cultures in test tubes supported a loss function in device 1. | |
| − | + | ||
| − | + | The Day 1 photos <b>(Fig.3)</b> show the three strains in order from strongest to weakest, with a negative control of regular <i>E. coli</i> on the right. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | The results of the lab generally followed our hypothesis. | + | |
| − | < | + | |
| − | < | + | |
| − | |||
| − | |||
| − | |||
Then, there are the Day 2 photos <b>(Fig.4)</b> in sets of three. Each of the three contains one of the three J23101 promoter containing device 1 cultures, with a J23106 promoter containing device 2 culture, a J23117 promoter containing device 3 culture, and the negative control, TOP 10, in that order. On day 2 the three J23101, device 1 cultures all looked different; one was still bright, one was entirely broken, and one was only half broken. | Then, there are the Day 2 photos <b>(Fig.4)</b> in sets of three. Each of the three contains one of the three J23101 promoter containing device 1 cultures, with a J23106 promoter containing device 2 culture, a J23117 promoter containing device 3 culture, and the negative control, TOP 10, in that order. On day 2 the three J23101, device 1 cultures all looked different; one was still bright, one was entirely broken, and one was only half broken. | ||
| − | |||
| − | |||
| − | |||
| − | < | + | Finally, the day 3 photos <b>(Fig.5)</b> show each set of three cultures along with one that has all three in order from strongest to weakest, with each set having the negative control at the end. |
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| + | {| cellpadding="2" | ||
| + | |- | ||
| + | | [[File:2015_Austin_UTexas_Interlab_ALLGRAPHS.png|340px|thumb|left|<b>Fig.2:</b> Average fluorescence from the plate reader of all biological replicates for each different Interlab study device over the course of three days, including TOP10 cells.]] | ||
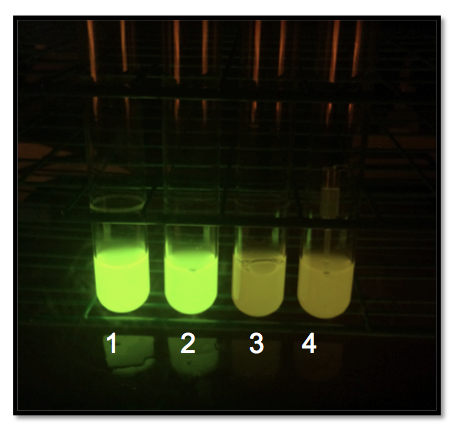
| + | | [[Image:2015_Austin_UTexas_Interlab_Day1.png|275px|thumb|left|<b>Fig.3:</b> Devices in cultures in our study as seen on Day 1. Device 4 is a negative control of TOP10 cells only.]] | ||
| + | <br style="clear:both" /> | ||
| + | |- | ||
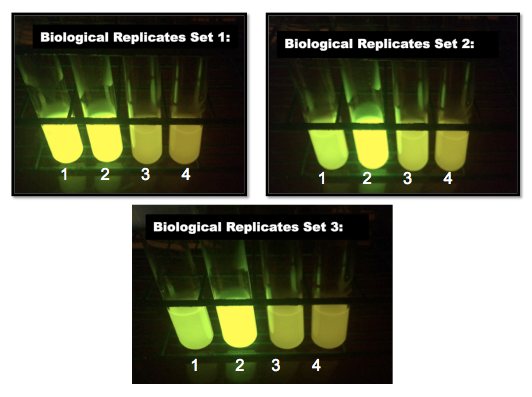
| + | | [[Image:2015_Austin_UTexas_Interlab_Day2.png|375px|thumb|right|<b>Fig.4:</b> Day 2 cultures of devices 1-3. <b>Set 1:</b> Tube 1: J23101+GFP Replicate A. Tube 2: J23106 + GFP Replicate A.Tube 3: J23117 + GFP Replicate A. Tube 4: TOP10 Cells <b>Set 2:</b> Order as described in Set 1, but for B replicates. <b>Set 3:</b> As in Set 1, but for C replicates.]] | ||
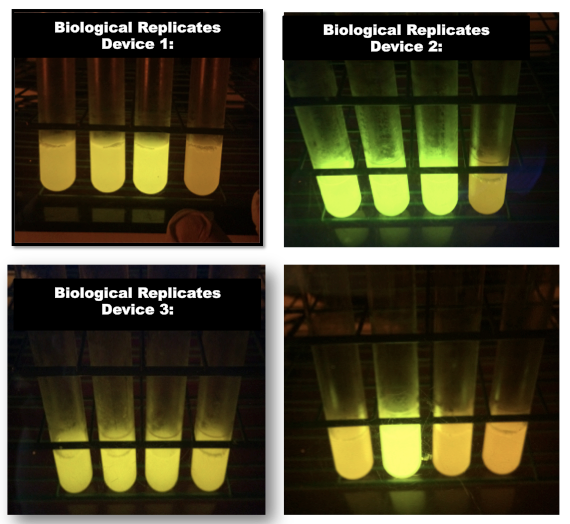
| + | | [[Image:2015_Austin_UTexas_Interlab_Day3.png|300px|thumb|right|<b>Fig.5:</b> (Cultures with Devices ordered 1, 2, 3, 4, from left to right). <b>Device 1:</b> From left to right: Biological Replicates A-C of J23101+GFP (Device 1) and TOP10 cells control on Day 3. <b>Device 2:</b> From left to right: Same as for "Device 1" but for Device 2, J23106+GFP. <b>Device 3:</b> From left to right: Same as for "Device 1", but for Device 3, J23117+GFP. <b>Bottom Right:</b> From left to right: Device 1, Device 2, Device 3, TOP10 cells control]] | ||
| + | |} | ||
| + | ===References=== | ||
| + | <div align="left">Anderson, C., Berkley iGEM Team., (2006). Anderson Promoter Collection. Registry of Standard Biological Parts. http://parts.igem.org/Promoters/Catalog/Anderson [accessed 19/09/2014] </div> | ||
{{Austin_UTexas_Footer}} | {{Austin_UTexas_Footer}} | ||
Latest revision as of 03:45, 19 September 2015
Interlab Measurement Study
The Interlab Study is an initiative started in 2014 by the iGEM foundation to collect standard measurement data from around the world. This year, as a part of the measurement track, we completed the Interlab study according to the directions provided on the iGEM Interlab Study webpage. The purpose of this year's study was to create three different GFP containing devices through BioBrick assembly and measure the fluorescence across biological replicates. Below, we have detailed our procedures, implications of our results, and conclusions we have drawn that relate back to our main focus of evolutionary stability in synthetic biology. In synthetic biology, it is important that part characterization is consistent between different labs to create well-defined standard parts for the Registry. We are excited to contribute to this study!
Protocol
1. Transform the genetic parts from the BioBrick 2015 kit into TOP10 E. coli cells. BioBrick assembly was used to engineer the InterLab study devices. Refer to the BioBrick website for more information on assembly procedure: http://parts.igem.org/Assembly:Standard_assembly.
2. Streak out an agar plate (LB/CAM) with the E. coli containing the device and any controls
- streak out one plate/device and control
- incubate plates overnight (18-20 hours) at 37°C
- “negative or background” Control: Cell background control (TOP10 cells)
- Negative Control: LB only control
Inoculate liquid culture with experimental devices (in triplicate) and controls in test tube in 10 ml LB/CAM media
Incubate liquid cultures for 16-18 hours at 300 rpm in 37°C
Plate Reader Measurement Protocol:
- Obtain initial OD600 measurement of O/N cultures
- Dilute the samples to an OD600 of 0.5
- calculate the dilution needed for each sample
- dilute each sample
- Re-measure your sample on OD600 to make sure it is at least within 5% of 0.5
Measure your samples
- add 80ul of each sample to the wells of a clear-bottomed 96 well plate
- The 96-well plate was inserted into an Infinite 200 PRO Microplate Reader
- Then, using the Tecan i-control the proper settings were selected and recorded.
- The Settings were: Excitation at 480 nm (9 nm width) and Emission at 525 nm (20 nm width) with optimal gain.
Calculate standard deviation and mean across triplicates of each device
Flow Cytometry Protocol:
- Wash culture in PBS to remove LB
- Load 150µl of sample into plate
- Prep the flow cytometer to read samples
- Prepare graphs
Extra Credit:
Three technical replicates of the experiment were recorded where the measurements were repeated. The experimental samples were measured at least three times with both the plate reader and flow cytometer. Furthermore, we conducted three days of measurements to observe any possible genetic breaking of the devices to confirm our hypothesis that the strongest promoter will confer the most metabolic load on the organism, which can cause loss-of-function mutations within the plasmid.
Protocol for further measurement of biological triplicates and technical replicates over the course of three days:
Day 0:
- Streak out frozen stock on plate
- Use the inoculation loop to streak out your cells onto LB/CAM plate.
- Incubate your plate at 37°C overnight for at least 20 hours.
Day 1:
- Pick 3 independent colonies and start three 5 ml O/N cultures in LB/CAM media.
- Grow overnight at 37°C and at 200-225RPM
Day 2:
- Start fresh 1:1000 dilution 5 ml cultures for each O/N culture.
- vortex the culture to generate an homogenous mixture
- Remove 250 µl from each O/N culture
- Use 5 µl of this for your fresh O/N culture
- Measure fluorescence intensity for your cultures with Flow Cytometer
- From the remaining culture in the Eppendorf, add 200 µl to a well within our 96 well plate.
Day 3:
- Repeat the procedure for Day 2
- Record the fluorescence readings and Flow Cytometer from the previous day
Devices Measured
1. The first device was cloned in lab and contains the following BioBrick parts:
- A promoter, BBa_J23101, a downstream gene (BBa_I13504), and a backbone (pSB1C3).
- The downstream gene is also a composite of a number of BioBrick parts including an RBS - B0034, a GFP coding sequence - E0040, and a transcriptional terminator - B0015.
2. The second device was cloned in lab and contains the following BioBrick parts:
- A promoter, BBa_J23106, a downstream gene (BBa_I13504), and a backbone (pSB1C3).
- The downstream gene is also a composite of a number of BioBrick parts including an RBS - B0034, a GFP coding sequence - E0040, and a transcriptional terminator - B0015.
3. The third device was cloned in lab and contains the following BioBrick parts:
- A promoter, BBa_J23117, a downstream gene (BBa_I13504), and a backbone (pSB1C3).
- The downstream gene is also a composite of a number of BioBrick parts including an RBS - B0034, a GFP coding sequence - E0040, and a transcriptional terminator - B0015.
Data and Discussion
The 2015 Interlab study provided a substantive way to continue our study of evolutionary stability in synthetic organisms. The framework of the study allowed us to understand the differences in metabolic load conferred by varying promoter strengths. All three devices created for this study were composed of the same downstream gene (BBa_I13504), and a backbone (pSB1C3). The order of promoter strength was as follows: J213101, J23106, J23117. The promoters used for the construction of the three devices are constitutive promoters of varying strengths, and should hypothetically generate varying metabolic load on the organism. All devices have the same coding sequence, RBS, transcriptional terminator, backbone, and thus plasmid copy number. We used the Interlab study as means to further our research to continue to measure this fitness cost imposed by identifying any point mutations, small insertions and deletions, large deletions, repeated operator sequences within the promoter, and insertion sequence (IS) element insertions in the sequencing data. It could help the effort to standardize promoter measurements between promoters in the same family, and help strengthen any evolutionary stability arguments made in the Breaking is Bad project.
The results of the lab generally followed our hypothesis. As seen from (Fig. 1 and 2), the J23101 promoter containing plasmid, device 1, with the strongest constitutive expression showed a significant loss in fluorescence after day 1, while the J23106 and J23117 promoter containing plasmids, device 2 and 3, respectively, were able to maintain stable levels of fluorescence over the course of three days. However, we expect these plasmids to eventually break. Although we did not sequence the broken cultures, visual assessment of the plates and cultures in test tubes supported a loss function in device 1.
The Day 1 photos (Fig.3) show the three strains in order from strongest to weakest, with a negative control of regular E. coli on the right.
Then, there are the Day 2 photos (Fig.4) in sets of three. Each of the three contains one of the three J23101 promoter containing device 1 cultures, with a J23106 promoter containing device 2 culture, a J23117 promoter containing device 3 culture, and the negative control, TOP 10, in that order. On day 2 the three J23101, device 1 cultures all looked different; one was still bright, one was entirely broken, and one was only half broken.
Finally, the day 3 photos (Fig.5) show each set of three cultures along with one that has all three in order from strongest to weakest, with each set having the negative control at the end.