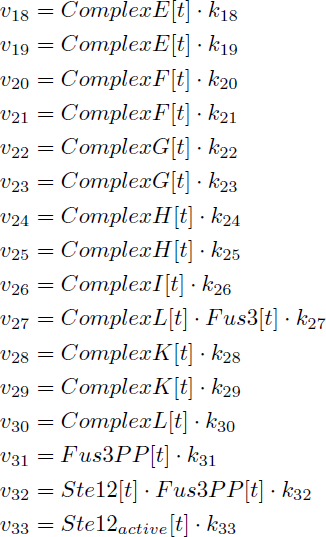Difference between revisions of "Team:Chalmers-Gothenburg/Detection"
SuperDavid (Talk | contribs) |
SuperDavid (Talk | contribs) |
||
| Line 23: | Line 23: | ||
[[File:ChalmersGothenburgModellingDas1.png|450px]]<br> | [[File:ChalmersGothenburgModellingDas1.png|450px]]<br> | ||
| − | <p class=figuretext> Figure 1. The modelling for the activation of the GPC receptor <p> | + | <p class=figuretext> Figure 1. The modelling for the activation of the GPC receptor </p> |
[[File:ChalmersGothenburgModellingDas2.png|450px]]<br> | [[File:ChalmersGothenburgModellingDas2.png|450px]]<br> | ||
| − | <p class=figuretext> Figure 2. The modelling for the activation of the G protein <p> | + | <p class=figuretext> Figure 2. The modelling for the activation of the G protein </p> |
[[File:ChalmersGothenburgModellingDas3.png|450px]]<br> | [[File:ChalmersGothenburgModellingDas3.png|450px]]<br> | ||
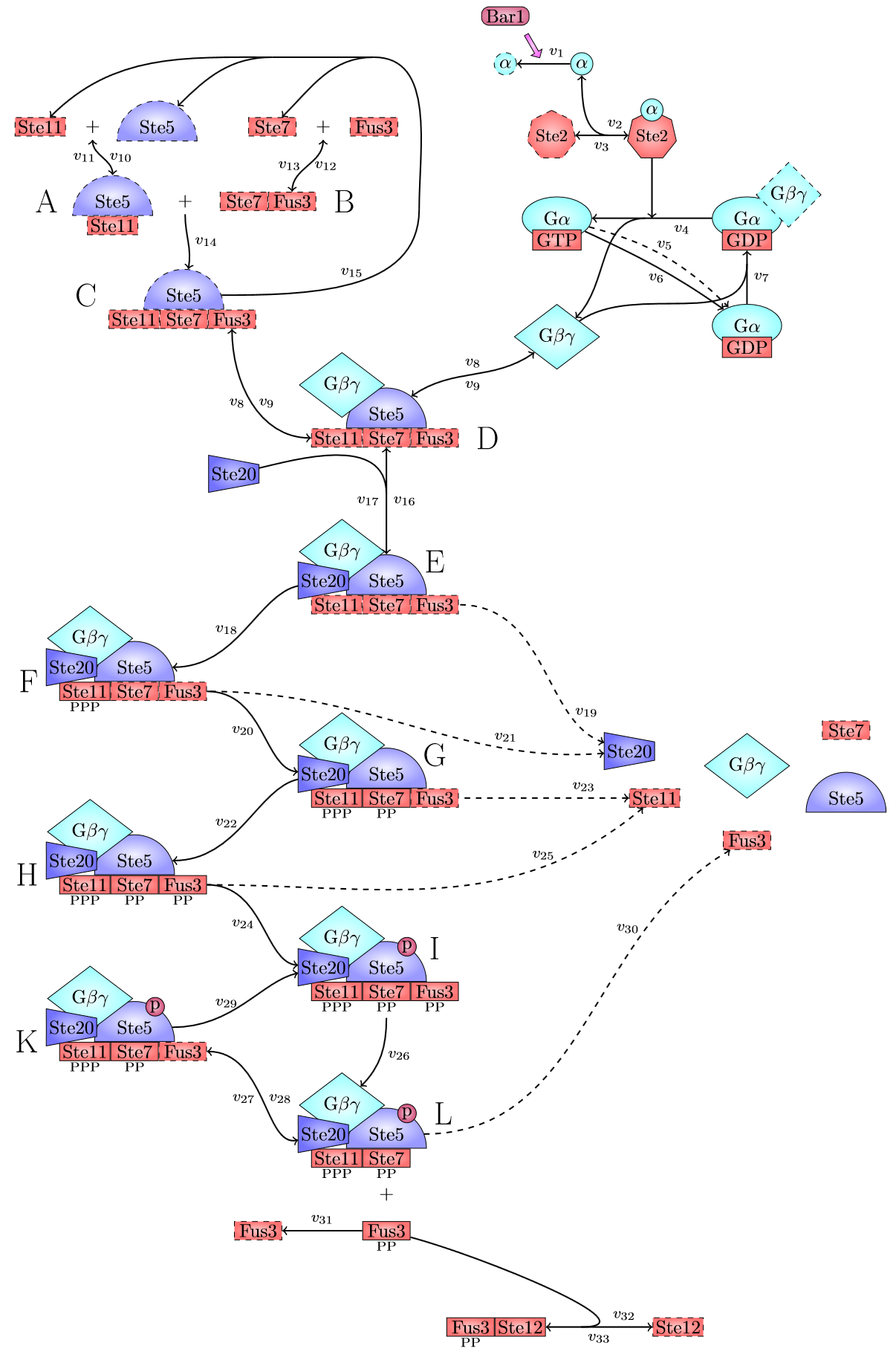
| − | <p class=figuretext> Figure 3. The modelling for the creation of the Ste5/Ste11/Ste7/Fus3 complex <p> | + | <p class=figuretext> Figure 3. The modelling for the creation of the Ste5/Ste11/Ste7/Fus3 complex </p> |
[[File:ChalmersGothenburgModellingDas5.png|450px]]<br> | [[File:ChalmersGothenburgModellingDas5.png|450px]]<br> | ||
| − | <p class=figuretext> Figure 4. The modelling for the activation of the Fus3 in the Ste5/Ste11/Ste7/Fus3 complex <p> | + | <p class=figuretext> Figure 4. The modelling for the activation of the Fus3 in the Ste5/Ste11/Ste7/Fus3 complex </p> |
[[File:ChalmersGothenburgModellingDas4.png|450px]]<br> | [[File:ChalmersGothenburgModellingDas4.png|450px]]<br> | ||
| − | <p class=figuretext> Figure 5. The modelling for part of the phosphorylation cascade in the Ste5/Ste11/Ste7/Fus3 complex<p> | + | <p class=figuretext> Figure 5. The modelling for part of the phosphorylation cascade in the Ste5/Ste11/Ste7/Fus3 complex</p> |
[[File:ChalmersGothenburgModellingDasKaskad.png|450px]] | [[File:ChalmersGothenburgModellingDasKaskad.png|450px]] | ||
| − | <p class=figuretext> Figure 6. The modelling for the phosphorylation cascade in the Ste5/Ste11/Ste7/Fus3 complex<p> | + | <p class=figuretext> Figure 6. The modelling for the phosphorylation cascade in the Ste5/Ste11/Ste7/Fus3 complex</p> |
[[File:ChalmersGothenburgModellingDas6.png|450px]]<br> | [[File:ChalmersGothenburgModellingDas6.png|450px]]<br> | ||
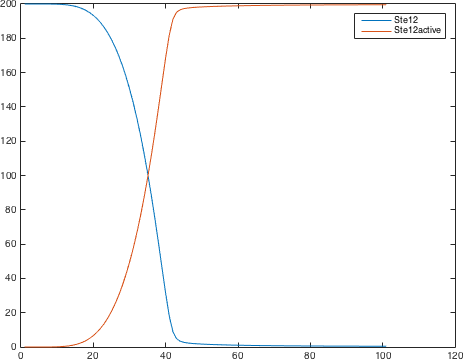
| − | <p class=figuretext> Figure 7. The modelling for the activation of Ste12<p> | + | <p class=figuretext> Figure 7. The modelling for the activation of Ste12</p> |
[[File:ChalmersGothenburgModellingDas7.png|450px]]<br> | [[File:ChalmersGothenburgModellingDas7.png|450px]]<br> | ||
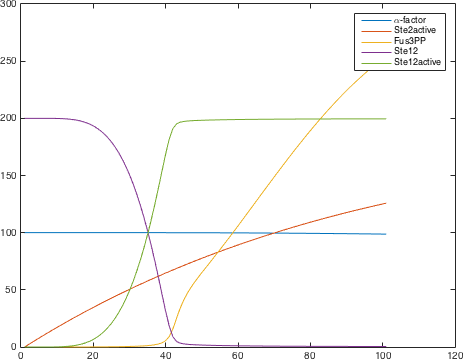
| − | <p class=figuretext> Figure 8. The modelling for the complete system, from activation of GPC receptor to activation of Ste12<p> | + | <p class=figuretext> Figure 8. The modelling for the complete system, from activation of GPC receptor to activation of Ste12</p> |
Revision as of 03:56, 19 September 2015


Detection
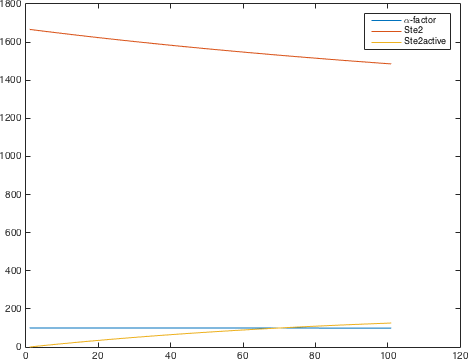
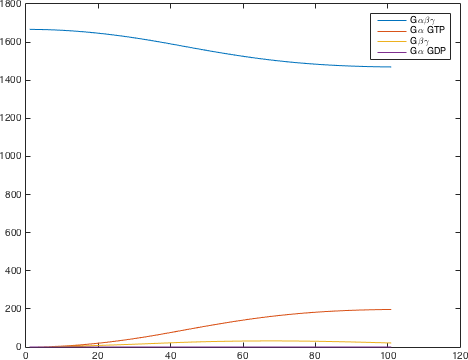
The detection system is used so allow Saccharomyces cerevisiae to detect a foreign peptide. The system couples a signaling pathway, the pheromone pathway with a receptor in from a species from a different another Subphylum. The pheromone pathway starts with a ligand receptor association followed by disassociation of g proteins associated to the receptor. The disassociated G protein starts a phosphorylation cascade (MAPK) leading to a activation of a transcriptionfactor [1].
Model Formation
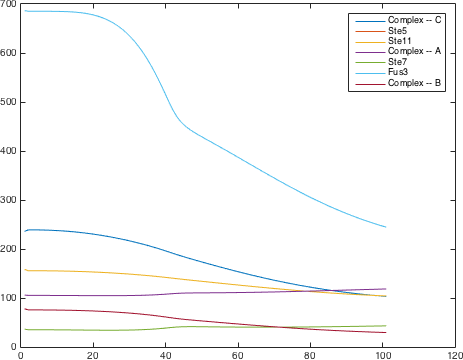
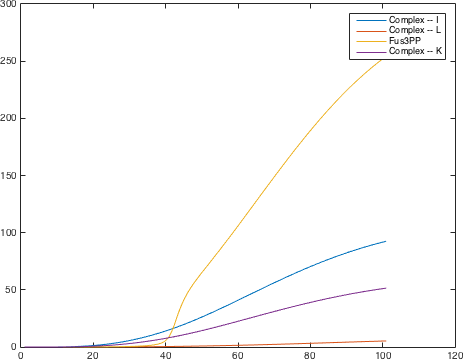
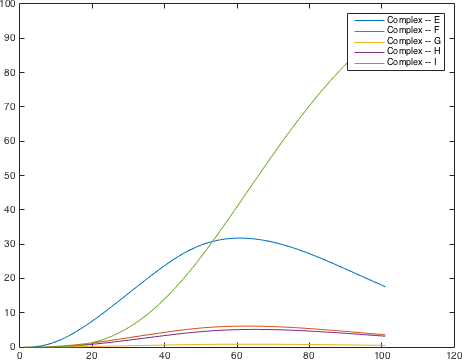
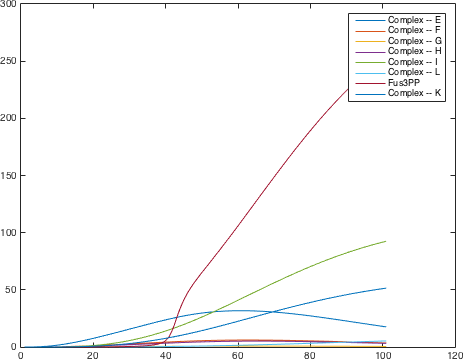
There are significant work on modeling the pheromone pathway, there is even a database dedicated to yeast pheromone pathway modeling (http://yeastpheromonemodel.org/). The receptor is a G protein coupled receptor (GPCR), the G proteins are the Gα, Gβ, and Gγ, the Gβ and Gγ proteins interactions are neglected in the model and the proteins are assumed to always be associated. A similar assumption is made on the Gα protein and its associated GDP, however, the GDP can still me phosphorylated to GTP. All the monomers present in the MAPK-cascade complexes were assumed not to be associated all the time. Also all MAPK-cascade were assumed to irreversible reactions except from the last reaction where Fus3 can re associate with the complex and then reenter the cascade. The activation Ste12 by the inhibition of Dig1 and Dig2 is a one-step reaction between Fus3pp and inactive Ste12. The Protein expression were assumed to be linear to the amount of active transcription factor.
The reactions was defines as:
Yielding the following differential equations:
Figure 1. The modelling for the activation of the GPC receptor
Figure 2. The modelling for the activation of the G protein
Figure 3. The modelling for the creation of the Ste5/Ste11/Ste7/Fus3 complex
Figure 4. The modelling for the activation of the Fus3 in the Ste5/Ste11/Ste7/Fus3 complex
Figure 5. The modelling for part of the phosphorylation cascade in the Ste5/Ste11/Ste7/Fus3 complex
Figure 6. The modelling for the phosphorylation cascade in the Ste5/Ste11/Ste7/Fus3 complex
Figure 7. The modelling for the activation of Ste12
Figure 8. The modelling for the complete system, from activation of GPC receptor to activation of Ste12
Parameters
| [Ste5] | 158.33 nM |
| [Ste11] | 158.33 nM |
| [Ste7] | 36.40 nM |
| [Fus3] | 686.40 nM |
| [Ste12] | 200 nM |
| [Ste20] | 1000 nM |
| [Gαβγ] | 1666.67 nM |
| [Ste5/Ste11] | 105.94 nM |
| [Ste7/Fus3] | 77.87 nM |
| [Ste5/Ste11/Ste7/Fus3] | 235.72 nM |
| Rest | 0 |
| k1 | 0.03 min−1 nM−1 | [1],[2] |
| k2 | 0.0012 min−1 nM−1 | [1],[2] |
| k3 | 0.6 min−1 | [1],[2] |
| k4 | 0.24 min−1 | [1],[2] |
| k5 | 0.024 min−1 | [1],[2] |
| k6 | 0.0036 min−1 nM−1 | [1],[2] |
| k7 | 0.24 min−1 | [1],[2] |
| k8 | 0.33 min−1 nM−1 | [1],[2] |
| k9 | 2000 min−1 nM−1 | [1],[2] |
| k10 | 0.1 min−1 nM−1 | [1] |
| k11 | 5 min−1 | [1] |
| k12 | 1 min−1 nM−1 | [1] |
| k13 | 3 min−1 | [1] |
| k14 | 1 min−1 nM−1 | [1],[3] |
| k15 | 3 min−1 | [1],[3] |
| k16 | 3 min−1 nM−1 | [1] |
| k17 | 100 min−1 | [1] |
| k18 | 5 min−1 nM−1 | [1] |
| k19 | 1 min−1 | [1] |
| k20 | 10 min−1 | [1] |
| k21 | 5 min−1 | [1] |
| k22 | 47 min−1 | [1],[4] |
| k23 | 5 min−1 | [1],[4] |
| k24 | 345 min−1 | [1],[4] |
| k25 | 5 min−1 | [1] |
| k26 | 50 min−1 | [1] |
| k27 | 5 min−1 | [1] |
| k28 | 140 min−1 | [1],[5] |
| k29 | 10 min−1 nM−1 | [1] |
| k30 | 1 min−1 | [1] |
| k31 | 250 min−1 | [1] |
| k32 | 5 min−1 | [1],[5] |
| k33 | 50 min−1 | [1] |
| k34 | 18 min−1 nM−1 | [1,[6]] |
| k35 | 10 min−1 | [1] |
References
[1] Kofahl, B., & Klipp, E. (2004). Modelling the dynamics of the yeast pheromone pathway. Yeast, 21(10), 831-850.
[2] Yi TM, Kitano H, Simon MI. 2003. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc Natl Acad Sci USA 100: 10 764–10 769
[3]Bardwell L, Cook JG Chang EC, Cairns BR, Thorner J. 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol 16: 3637–3650
[4] Ferrell JE Jr, Bhatt RR. 1997. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J Biol Chem 272: 19 008–19 016.
[5] van Drogen F, Stucke VM, Jorritsma G, Peter M. 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nature Cell Biol 3: 1051–1059
[6] Prowse CN, Deal MS, Lew J. 2001. The complete pathway for catalytic activation of the mitogen-activated protein kinase, ERK2. J Biol Chem 276: 40 817–40 823.