Difference between revisions of "Team:Birkbeck/Composite Part"
| (16 intermediate revisions by the same user not shown) | |||
| Line 12: | Line 12: | ||
<p>The tfa (tail fibre assembly) protein of bacteriophage lambda assists in the assembly of the stf (short tail fibre) protein into a functional short tail fibre [1],[2]. Tfa gene displays a high level of homology (~40%) with the gp38 of bacteriophage T4 [2],[3],[4]. | <p>The tfa (tail fibre assembly) protein of bacteriophage lambda assists in the assembly of the stf (short tail fibre) protein into a functional short tail fibre [1],[2]. Tfa gene displays a high level of homology (~40%) with the gp38 of bacteriophage T4 [2],[3],[4]. | ||
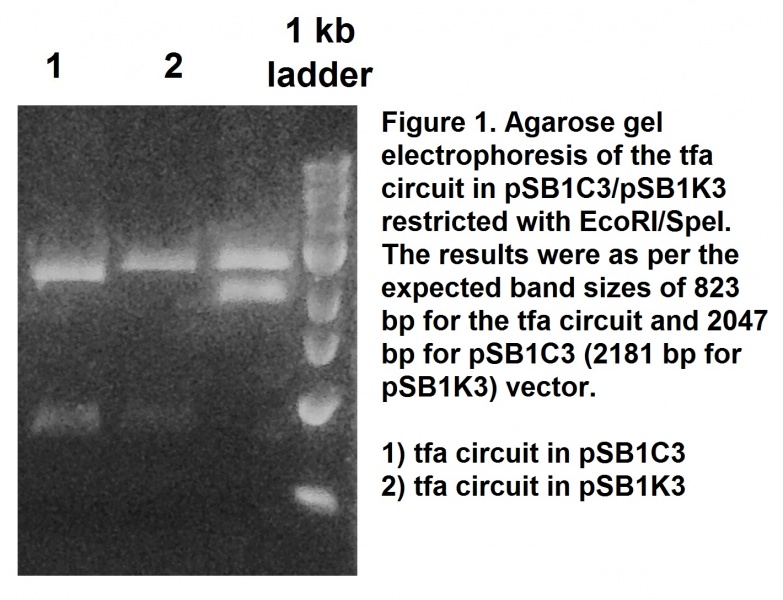
| − | <p>This tfa circuit was synthesised removing the illegal restriction sites to make the sequence BioBrick-compatible. The sequence was cloned into a pSB1C3 and the success of the cloning procedure was confirmed by restriction with EcoRI and SpeI restriction enzymes and followed by agarose gel electrophoresis (Figure 1). The construct was also confirmed by sequencing. This BioBrick was registered as part <a href="http://parts.igem.org/Part:BBa_K1846001">BBa_K1846001</a>.</p> | + | <p>This tfa circuit was synthesised removing the illegal restriction sites to make the sequence BioBrick-compatible. The sequence was cloned into a pSB1C3 and the success of the cloning procedure was confirmed by restriction with <i>EcoRI</i> and <i>SpeI</i> restriction enzymes and followed by agarose gel electrophoresis (Figure 1). The construct was also confirmed by sequencing. This BioBrick was registered as part <a href="http://parts.igem.org/Part:BBa_K1846001">BBa_K1846001</a>.</p> |
| − | <p><img alt="File:BBKiGEM-tfa-circuit.jpg" src="/wiki/images/thumb/f/f3/BBKiGEM-tfa-circuit.jpg/783px-BBKiGEM-tfa-circuit.jpg" width=" | + | <p><img alt="File:BBKiGEM-tfa-circuit.jpg" src="/wiki/images/thumb/f/f3/BBKiGEM-tfa-circuit.jpg/783px-BBKiGEM-tfa-circuit.jpg" width="350" height="269" srcset="/wiki/images/thumb/f/f3/BBKiGEM-tfa-circuit.jpg/1175px-BBKiGEM-tfa-circuit.jpg 1.5x, /wiki/images/f/f3/BBKiGEM-tfa-circuit.jpg 2x"></p> |
<br> | <br> | ||
<br> | <br> | ||
| Line 22: | Line 22: | ||
<IMG SRC="https://static.igem.org/mediawiki/2015/e/ef/Birkbeck_tetR_circuit.png" height=120 width=440> | <IMG SRC="https://static.igem.org/mediawiki/2015/e/ef/Birkbeck_tetR_circuit.png" height=120 width=440> | ||
| − | <p>This is a constitutive part for the production of the tetracyline repressor TetR, using a constitutive, medium-copy chloramphenicol promoter (<a href="http://parts.igem.org/Part:BBa_I14033">BBa_I14033</a>), ribosome binding sequence (<a href="http://parts.igem.org/Part:BBa_B0031">BBa_B0031</a>) and double terminator (rrnb T1 terminator followed by a T7Te terminator <a href="http://parts.igem.org/Part:BBa_B0010">BBa_B0010</a>).</p> | + | <p>This is a constitutive part for the production of the tetracyline repressor TetR, using a constitutive, medium-copy chloramphenicol promoter (<a href="http://parts.igem.org/Part:BBa_I14033">BBa_I14033</a>), ribosome binding sequence (<a href="http://parts.igem.org/Part:BBa_B0031">BBa_B0031</a>) and double terminator (rrnb T1 terminator followed by a T7Te terminator, <a href="http://parts.igem.org/Part:BBa_B0010">BBa_B0010</a>).</p> |
| − | + | ||
| − | + | ||
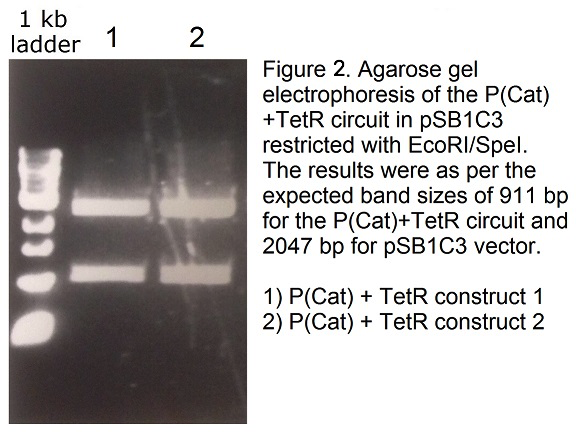
| + | <p>TetR generator cassette (<a href="http://parts.igem.org/Part:BBa_P0140">BBa_P0140</a>) and P(Cat) promoter (<a href="http://parts.igem.org/Part:BBa_I14033">BBa_I14033</a>) were obtained from 2015 iGEM distribution kit and cloned into a single pSB1C3 vector. Correct cloning of the parts into the pSB1C3 shipping vector was confirmed by agarose gel electrophoresis (Figure 2) and Sanger sequencing. This BioBrick has been registered as part <a href="http://parts.igem.org/Part:BBa_K1846003">BBa_K1846003</a>. | ||
| + | <img alt="File:BBKiGEM-Pcat-TetR2.jpg" src="/wiki/images/6/65/BBKiGEM-Pcat-TetR2.jpg" width="350" height="269"> | ||
| + | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 33: | Line 34: | ||
<h3><b>TetR-regulated tfa (tail fibre assembly) circuit (BBa_K1846007)</b></h3> | <h3><b>TetR-regulated tfa (tail fibre assembly) circuit (BBa_K1846007)</b></h3> | ||
<IMG SRC="https://static.igem.org/mediawiki/2015/c/c3/Birkbeck_tetR_tfa_circuit.png" height=120 width=900> | <IMG SRC="https://static.igem.org/mediawiki/2015/c/c3/Birkbeck_tetR_tfa_circuit.png" height=120 width=900> | ||
| − | <p> | + | <p>A circuit controlling the production of the tail fibre assembly (tfa) protein of bacteriophage lambda to prevent toxicity to the cell. The tfa gene operates under a TetR repressible promoter, while a second circuit (<a href="http://parts.igem.org/Part:BBa_K1846003">BBa_K1846003</a>) produces TetR (tetracycline repressor) under control of a P(Cat) promoter. With the production of the tfa protein under control of a Tet-repressible promoter, the tfa coding sequence will not be expressed due to P(TetR). However, on addition of anhydrous tetracycline (aTc), TetR will preferably bind to aTc molecule, alleviating repression of P(TetR) and allowing the initiation of transcription. The correct cloning of this BioBrick was confirmed through Sanger sequencing and agarose gel electrophoresis (Figure 3). |
| + | <p> | ||
| + | <img alt="File:BBKiGEM-P(Cat)+TetR+tfa-circuit.jpg" src="/wiki/images/0/0c/BBKiGEM-P%28Cat%29%2BTetR%2Btfa-circuit.jpg" width="400" height="380"></p> | ||
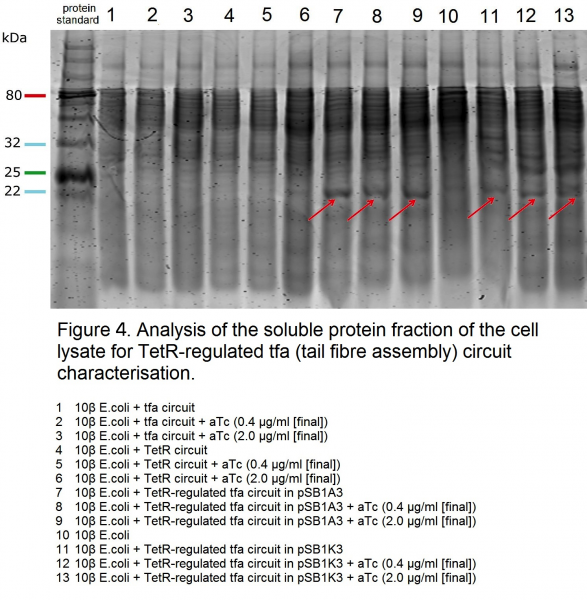
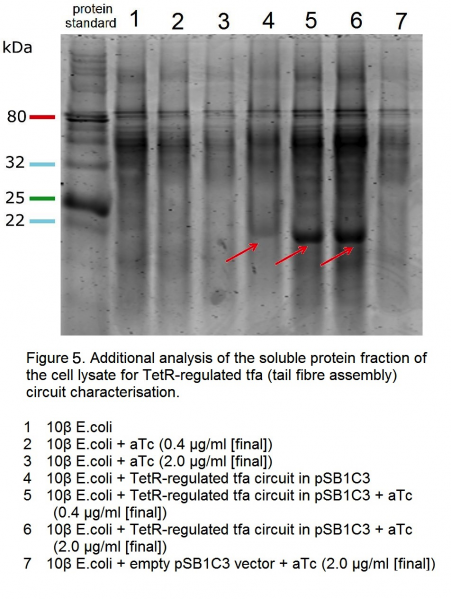
| − | <p>We characterised this construct by analysing the soluble protein fraction of the cell lysate ( | + | <p>We characterised this construct by analysing the soluble protein fraction of the cell lysate (Figures 4 and 5). Under the TetR regulation, <i>tfa</i> gene is expressed under a tight control, and can be visualised on the gel (tfa protein = 194 aa, MW: 21602.2 Da), (Figure 4, samples 7, 8, 9 and 11, 12 and 13; Figure 5, samples 4, 5 and 6). <i>tfa</i> gene circuit on its own does not appear to produce tfa protein bands (Figure 4, samples 1, 2 and 3). Although the literature suggests that the protein is soluble even with high expression [1], we hypothesise the lack of bands in these samples is due to the protein being highly acidic, and thus toxic to the cell. It may be that due to the toxicity the cells that express less of the protein are selected for in these experimental conditions. Under the regulation of TetR, even when induced, the expression of the <i>tfa</i> gene is considerably lower than when unregulated and hence, we hypothesise, a considerable amount of soluble material can be recovered. The TetR repressor contains an LVA tag for rapid degradation and hence in the uninduced samples 7 and 11 in Figure 4, and sample 4 in Figure 5, tfa protein bands can still be observed.</p> |
| − | < | + | |
| − | < | + | <img alt="File:BBKiGEM-SDSgel-Figure 1b.png" src="/wiki/images/thumb/1/1d/BBKiGEM-SDSgel-Figure_1b.png/587px-BBKiGEM-SDSgel-Figure_1b.png" width="455" height="465" srcset="/wiki/images/thumb/1/1d/BBKiGEM-SDSgel-Figure_1b.png/881px-BBKiGEM-SDSgel-Figure_1b.png 1.5x, /wiki/images/thumb/1/1d/BBKiGEM-SDSgel-Figure_1b.png/1174px-BBKiGEM-SDSgel-Figure_1b.png 2x"> |
| + | |||
| + | <img alt="File:BBKiGEM-SDSgel-Figure 2b.png" src="/wiki/images/thumb/5/59/BBKiGEM-SDSgel-Figure_2b.png/451px-BBKiGEM-SDSgel-Figure_2b.png" width="350" height="465" srcset="/wiki/images/thumb/5/59/BBKiGEM-SDSgel-Figure_2b.png/677px-BBKiGEM-SDSgel-Figure_2b.png 1.5x, /wiki/images/thumb/5/59/BBKiGEM-SDSgel-Figure_2b.png/902px-BBKiGEM-SDSgel-Figure_2b.png 2x"> | ||
| + | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 44: | Line 50: | ||
<h3><b>cI-Cro circuit (BBa_K1846005)</b></h3> | <h3><b>cI-Cro circuit (BBa_K1846005)</b></h3> | ||
<IMG SRC="https://static.igem.org/mediawiki/2015/2/28/Birkbeck_cI-cro_circuit.png" height=120 width=900> | <IMG SRC="https://static.igem.org/mediawiki/2015/2/28/Birkbeck_cI-cro_circuit.png" height=120 width=900> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | + | <p>The cI-Cro construct contains a circuit for the production - via a constitutive promoter - of the cI repressor protein. The circuit uses a T7 promoter to drive the expression of the <i>cro</i> gene in the opposite direction to the <i>cI</i> gene, essentially silencing the expression of the <i>cI</i> gene. The strength of the promoter used means that in the presence of T7 DNA polymerase, production of the Cro repressor protein will incapacitate the production of the cI repressor protein. Thus, the presence of the T7 RNA polymerase in the system enables the switch from the lysogenic cycle to the lytic cycle.</p> |
| − | < | + | <p>The cI repressor protein (also known as lambda repressor) is responsible for keeping bacteriophage lambda in the lysogenic cycle through the cooperative binding of two repressor dimers to the DNA, repressing the expression of the <i>cro</i> gene [5],[6]. Cro repressor protein, on the other hand, controls the late stage of the lytic development. When the concentration of the Cro repressor is greater than that of the cI repressor, the lytic cycle is initiated [5],[6]. |
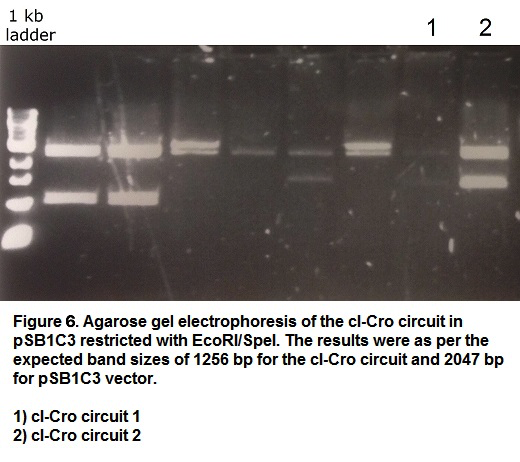
| + | <p>This cI-Cro circuit was synthesised removing the illegal restriction sites to make the sequence BioBrick-compatible. The sequence was cloned into a pSB1C3 and the success of the cloning procedure was confirmed by restriction with <i>EcoRI</i> and <i>SpeI</i> restriction enzymes and followed by agarose gel electrophoresis (Figure 6). The construct was also confirmed by sequencing. This BioBrick was registered as part <a href="http://parts.igem.org/Part:BBa_K1846005">BBa_K1846005</a>. | ||
| + | <p> | ||
| + | <img alt="File:BBKiGEM-cICro-circuit.png" src="/wiki/images/b/bd/BBKiGEM-cICro-circuit.png" width="350" height="311"> | ||
| + | |||
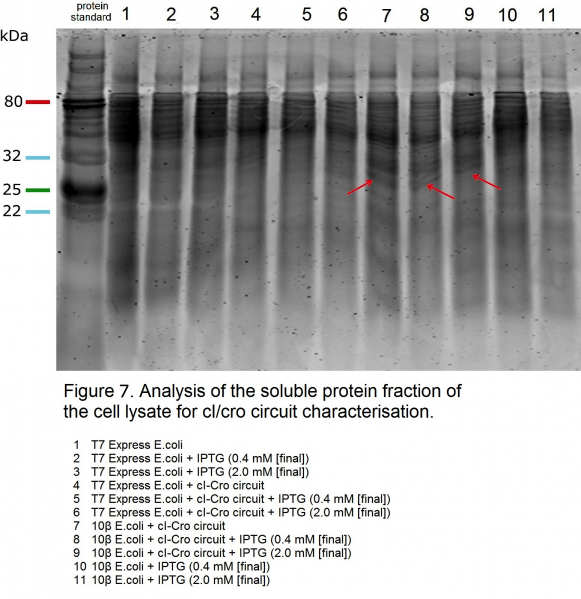
| + | <p>We characterised this construct by analysing the soluble protein fraction of the cell lysate (Figure 7). As cI repressor protein is produced constitutively in our circuit, we expected to see a cI repressor protein band at ca 26 kDa (cI repressor protein = 237 aa, MW: 26211.8 Da). Our control <i>E.coli</i> 10β cells containing the cI-Cro construct have shown the expected bands at ca 26 kDa (Figure 7, samples 7, 8 and 9). We attribute the lack of cI protein bands in our T7 Express cell lysate (Figure 7, sample 4) to T7 RNAP leakage, which would silence the expression of the <i>cI</i> gene through expression of <i>cro</i> gene in the opposite direction. Absence of Cro repressor protein bands (Cro repressor protein = 66 aa; MW: 7363.4 Da) could be due to protein overexpression and aggregation into insoluble fraction (due to the strength of the T7 promoter). This hypothesis requires further investigation, however our preliminary results suggest the circuit would be functional. | ||
| + | |||
| + | Previously existing BioBricks or parts containing either cI or Cro repressors include: <a href="http://parts.igem.org/Part:BBa_K150001">BBa_K150001</a>; <a href="http://parts.igem.org/Part:BBa_K150004">BBa_K150004</a>; <a href="http://parts.igem.org/Part:BBa_K1195004">BBa_K1195004</a>; <a href="http://parts.igem.org/Part:BBa_I741111">BBa_I741111</a>; <a href="http://parts.igem.org/Part:BBa_K177009">BBa_K177009</a>; <a href="http://parts.igem.org/Part:BBa_K648028">BBa_K648028</a>; <a href="http://parts.igem.org/Part:BBa_K648128">BBa_K648128</a>; <a href="http://parts.igem.org/Part:BBa_K648132">BBa_K648132</a>. | ||
| + | <p> | ||
| + | |||
| + | <img alt="File:BBKiGEM-SDSgel-Figure 3b.png" src="/wiki/images/thumb/d/d8/BBKiGEM-SDSgel-Figure_3b.png/581px-BBKiGEM-SDSgel-Figure_3b.png" width="455" height="465" srcset="/wiki/images/thumb/d/d8/BBKiGEM-SDSgel-Figure_3b.png/872px-BBKiGEM-SDSgel-Figure_3b.png 1.5x, /wiki/images/thumb/d/d8/BBKiGEM-SDSgel-Figure_3b.png/1162px-BBKiGEM-SDSgel-Figure_3b.png 2x"> | ||
| + | <p> | ||
<br> | <br> | ||
| Line 59: | Line 71: | ||
<p>[3] Hendrix, R. W., & Duda, R. L. (1992). Bacteriophage lambda PaPa: not the mother of all lambda phages. Science (New York, N.Y.), 258(5085), 1145–1148. doi:10.1126/science.1439823</p> | <p>[3] Hendrix, R. W., & Duda, R. L. (1992). Bacteriophage lambda PaPa: not the mother of all lambda phages. Science (New York, N.Y.), 258(5085), 1145–1148. doi:10.1126/science.1439823</p> | ||
<p>[4] Haggard-Ljungquist, E., Halling, C., & Calendar, R. (1992). DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. Journal of Bacteriology, 174(5), 1462–1477.</p> | <p>[4] Haggard-Ljungquist, E., Halling, C., & Calendar, R. (1992). DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. Journal of Bacteriology, 174(5), 1462–1477.</p> | ||
| + | <p>[5] Echols, H., & Murialdo, H. (1978). Genetic map of bacteriophage lambda. Microbiological Reviews, 42(3), 577–591.</p> | ||
| + | <p>[6] Friedman, D. I., & Court, D. L. (2001). Bacteriophage lambda: Alive and well and still doing its thing. Current Opinion in Microbiology, 4(2), 201–207. doi:10.1016/S1369-5274(00)00189-2</p> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <li><a href="https://2015.igem.org/Team:Birkbeck/BioBricks">Return to BioBricks overview</a></li> | ||
| + | <li><a href="https://2015.igem.org/Team:Birkbeck/Basic_Parts">Have a look at our Basic Parts</a></li> | ||
| + | |||
| + | </br> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 21:16, 16 November 2015
Our BioBricks
Composite Parts
tfa (tail fibre assembly) gene circuit (BBa_K1846001)

This part provides the gene sequence for tfa (tail fibre assembly) protein, together with a TetR repressible promoter (BBa_R0040), ribosome binding site (BBa_B0034) and an rrNB T1 terminator (BBa_B0010).
The tfa (tail fibre assembly) protein of bacteriophage lambda assists in the assembly of the stf (short tail fibre) protein into a functional short tail fibre [1],[2]. Tfa gene displays a high level of homology (~40%) with the gp38 of bacteriophage T4 [2],[3],[4].
This tfa circuit was synthesised removing the illegal restriction sites to make the sequence BioBrick-compatible. The sequence was cloned into a pSB1C3 and the success of the cloning procedure was confirmed by restriction with EcoRI and SpeI restriction enzymes and followed by agarose gel electrophoresis (Figure 1). The construct was also confirmed by sequencing. This BioBrick was registered as part BBa_K1846001.
TetR circuit (BBa_K1846003)

This is a constitutive part for the production of the tetracyline repressor TetR, using a constitutive, medium-copy chloramphenicol promoter (BBa_I14033), ribosome binding sequence (BBa_B0031) and double terminator (rrnb T1 terminator followed by a T7Te terminator, BBa_B0010).
TetR generator cassette (BBa_P0140) and P(Cat) promoter (BBa_I14033) were obtained from 2015 iGEM distribution kit and cloned into a single pSB1C3 vector. Correct cloning of the parts into the pSB1C3 shipping vector was confirmed by agarose gel electrophoresis (Figure 2) and Sanger sequencing. This BioBrick has been registered as part BBa_K1846003.
TetR-regulated tfa (tail fibre assembly) circuit (BBa_K1846007)

A circuit controlling the production of the tail fibre assembly (tfa) protein of bacteriophage lambda to prevent toxicity to the cell. The tfa gene operates under a TetR repressible promoter, while a second circuit (BBa_K1846003) produces TetR (tetracycline repressor) under control of a P(Cat) promoter. With the production of the tfa protein under control of a Tet-repressible promoter, the tfa coding sequence will not be expressed due to P(TetR). However, on addition of anhydrous tetracycline (aTc), TetR will preferably bind to aTc molecule, alleviating repression of P(TetR) and allowing the initiation of transcription. The correct cloning of this BioBrick was confirmed through Sanger sequencing and agarose gel electrophoresis (Figure 3).

We characterised this construct by analysing the soluble protein fraction of the cell lysate (Figures 4 and 5). Under the TetR regulation, tfa gene is expressed under a tight control, and can be visualised on the gel (tfa protein = 194 aa, MW: 21602.2 Da), (Figure 4, samples 7, 8, 9 and 11, 12 and 13; Figure 5, samples 4, 5 and 6). tfa gene circuit on its own does not appear to produce tfa protein bands (Figure 4, samples 1, 2 and 3). Although the literature suggests that the protein is soluble even with high expression [1], we hypothesise the lack of bands in these samples is due to the protein being highly acidic, and thus toxic to the cell. It may be that due to the toxicity the cells that express less of the protein are selected for in these experimental conditions. Under the regulation of TetR, even when induced, the expression of the tfa gene is considerably lower than when unregulated and hence, we hypothesise, a considerable amount of soluble material can be recovered. The TetR repressor contains an LVA tag for rapid degradation and hence in the uninduced samples 7 and 11 in Figure 4, and sample 4 in Figure 5, tfa protein bands can still be observed.
cI-Cro circuit (BBa_K1846005)

The cI-Cro construct contains a circuit for the production - via a constitutive promoter - of the cI repressor protein. The circuit uses a T7 promoter to drive the expression of the cro gene in the opposite direction to the cI gene, essentially silencing the expression of the cI gene. The strength of the promoter used means that in the presence of T7 DNA polymerase, production of the Cro repressor protein will incapacitate the production of the cI repressor protein. Thus, the presence of the T7 RNA polymerase in the system enables the switch from the lysogenic cycle to the lytic cycle.
The cI repressor protein (also known as lambda repressor) is responsible for keeping bacteriophage lambda in the lysogenic cycle through the cooperative binding of two repressor dimers to the DNA, repressing the expression of the cro gene [5],[6]. Cro repressor protein, on the other hand, controls the late stage of the lytic development. When the concentration of the Cro repressor is greater than that of the cI repressor, the lytic cycle is initiated [5],[6].
This cI-Cro circuit was synthesised removing the illegal restriction sites to make the sequence BioBrick-compatible. The sequence was cloned into a pSB1C3 and the success of the cloning procedure was confirmed by restriction with EcoRI and SpeI restriction enzymes and followed by agarose gel electrophoresis (Figure 6). The construct was also confirmed by sequencing. This BioBrick was registered as part BBa_K1846005.
We characterised this construct by analysing the soluble protein fraction of the cell lysate (Figure 7). As cI repressor protein is produced constitutively in our circuit, we expected to see a cI repressor protein band at ca 26 kDa (cI repressor protein = 237 aa, MW: 26211.8 Da). Our control E.coli 10β cells containing the cI-Cro construct have shown the expected bands at ca 26 kDa (Figure 7, samples 7, 8 and 9). We attribute the lack of cI protein bands in our T7 Express cell lysate (Figure 7, sample 4) to T7 RNAP leakage, which would silence the expression of the cI gene through expression of cro gene in the opposite direction. Absence of Cro repressor protein bands (Cro repressor protein = 66 aa; MW: 7363.4 Da) could be due to protein overexpression and aggregation into insoluble fraction (due to the strength of the T7 promoter). This hypothesis requires further investigation, however our preliminary results suggest the circuit would be functional. Previously existing BioBricks or parts containing either cI or Cro repressors include: BBa_K150001; BBa_K150004; BBa_K1195004; BBa_I741111; BBa_K177009; BBa_K648028; BBa_K648128; BBa_K648132.
[1] Hashemolhosseini, S., Stierhof, Y. D., Hindennach, I., & Henning, U. (1996). Characterization of the helper proteins for the assembly of tail fibers of coliphages T4 and λ. Journal of Bacteriology, 178(21), 6258–6265.
[2] Montag, D., Schwarz, H., & Henning, U. (1989). A component of the side tail fiber of E. coli bacteriophage λ can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. Journal of Bacteriology, 171(8), 4378–4384.
[3] Hendrix, R. W., & Duda, R. L. (1992). Bacteriophage lambda PaPa: not the mother of all lambda phages. Science (New York, N.Y.), 258(5085), 1145–1148. doi:10.1126/science.1439823
[4] Haggard-Ljungquist, E., Halling, C., & Calendar, R. (1992). DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. Journal of Bacteriology, 174(5), 1462–1477.
[5] Echols, H., & Murialdo, H. (1978). Genetic map of bacteriophage lambda. Microbiological Reviews, 42(3), 577–591.
[6] Friedman, D. I., & Court, D. L. (2001). Bacteriophage lambda: Alive and well and still doing its thing. Current Opinion in Microbiology, 4(2), 201–207. doi:10.1016/S1369-5274(00)00189-2




