Difference between revisions of "Team:Birkbeck/Basic Parts"
| (26 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
<h2>Basic Parts</h2> | <h2>Basic Parts</h2> | ||
| + | |||
<h3><b>ORF314 (BBa_K1846000)</b></h3> | <h3><b>ORF314 (BBa_K1846000)</b></h3> | ||
<img src="https://static.igem.org/mediawiki/2015/5/5a/Birkbeck_ORF314_phage.png"> | <img src="https://static.igem.org/mediawiki/2015/5/5a/Birkbeck_ORF314_phage.png"> | ||
| − | <p> | + | <p>This is host-specific subunit of the short tail fibre protein of bacteriophage lambda. The C-terminus of the protein (ORF-314) contains the recognition site that confers binding specificity to the OmpC membrane protein of <i>E.coli</i>[1].</p> |
| − | <p>To transform | + | <p>To transform ORF-314 sequence into a BioBrick basic part, we had the coding sequence synthesised, including the BioBrick prefix and suffix. The sequence was optimised to remove illegal restriction sites. We restricted both our synthesised ORF-314 and the linearised plasmid backbones (pSB1C3 for shipping, and pSB1K3 for further processing) with <i>EcoRI</i> and <i>PstI</i> restriction enzymes, before ligating the two pieces together using T4 DNA ligase. Success of the cloning procedure was confirmed by restriction with <i>EcoRI</i> and <i>PstI</i> restriction enzymes followed by agarose gel electrophoresis (Figure 1), with pSB1C3 alone and ORF314 alone as controls. The construct was also confirmed by sequencing. ORF-314 was submitted as <a href="http://parts.igem.org/Part:BBa_K1846000">BBa_K1846000.</a></p> |
| − | + | <a href='http://postimage.org/' target='_blank'><img src='http://s23.postimg.org/8de2ulxyz/150730_ORF314_p_SB1_C3_annotated.jpg' border='0' alt="Agarose gel electrophoresis of ORF314 in vector" /></a><br /><a target='_blank' href='http://postimage.org/'></a><p> | |
| − | <a href='http://postimage.org/' target='_blank'><img src='http://s23.postimg.org/8de2ulxyz/150730_ORF314_p_SB1_C3_annotated.jpg' border='0' alt="Agarose gel electrophoresis of ORF314 in vector" /></a><br /><a target='_blank' href='http://postimage.org/'></a>< | + | <p><b>Figure 1. Agarose gel electrophoresis of ORF314 inserted in pSB1C3 and restricted with <i>EcoRI</i> and <i>PstI</i>. The expected band sizes are 1 kb for ORF314 and 2 kb for pSB1C3. Successful results were obtained for samples 2 and 3.</p></b> |
| − | <p><b> | + | |
<br> | <br> | ||
| Line 18: | Line 18: | ||
<img src="https://static.igem.org/mediawiki/2015/f/fd/Birkbeck_stf_phage.png"> | <img src="https://static.igem.org/mediawiki/2015/f/fd/Birkbeck_stf_phage.png"> | ||
<p>The <i>stf</i> gene encodes the short tail fibres found in the Ur-lambda bacteriophage, the original isolate [1]. The wild type lambda strain (λ-PaPa) that is most commonly used in research carries a frameshift mutation in the <i>stf</i> gene sequence producing no such fibres. It has been shown that the presence of these additional thin tails results in the more efficient adsorption to the host, <i>E.coli</i> [1]. | <p>The <i>stf</i> gene encodes the short tail fibres found in the Ur-lambda bacteriophage, the original isolate [1]. The wild type lambda strain (λ-PaPa) that is most commonly used in research carries a frameshift mutation in the <i>stf</i> gene sequence producing no such fibres. It has been shown that the presence of these additional thin tails results in the more efficient adsorption to the host, <i>E.coli</i> [1]. | ||
| − | The <i>stf</i> gene is a product of two open reading frames: ORF-401 and ORF-314. Introduction of 1 bp just after ORF-401 removes the frameshift fusing the two coding regions together which results in a functional short tail fibre protein consisting of 774 aa [2]. The C-terminal end of the stf protein (i.e. ORF-314) offers host receptor recognition for OmpC (Outer membrane protein C) on the surface of <i>E.coli</i>, widening receptor specificity and host range relative to λ-PaPa which only recognises the <i>E.coli</i> Maltoporin (<i>lamB</i> gene product) through its tip attachment protein J [1],[3]. Additionally, the C-terminus (ORF-314) displays a high level of homology with the gp37 of bacteriophage T4 [4]. This BioBrick was registered as part <a href="http://parts.igem.org/Part:BBa_K1846004">BBa_K1846004</a>.</p> | + | <p>The <i>stf</i> gene is a product of two open reading frames: ORF-401 and ORF-314. Introduction of 1 bp just after ORF-401 removes the frameshift fusing the two coding regions together which results in a functional short tail fibre protein consisting of 774 aa [2]. The C-terminal end of the stf protein (i.e. ORF-314) offers host receptor recognition for OmpC (Outer membrane protein C) on the surface of <i>E.coli</i>, widening receptor specificity and host range relative to λ-PaPa which only recognises the <i>E.coli</i> Maltoporin (<i>lamB</i> gene product) through its tip attachment protein J [1],[3]. Additionally, the C-terminus (ORF-314) displays a high level of homology with the gp37 of bacteriophage T4 [4].</p> |
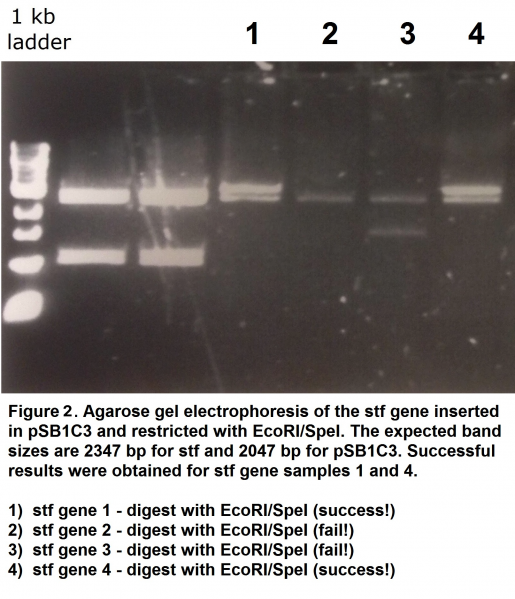
| + | <p>The gene has been assembled using the synthesised ORF-401 with a linker sequence (removing the frameshift mutation) and ORF-314. The sequences were optimised to remove illegal restriction sites to make the sequence BioBrick-compatible. We cloned the two open reading frames and a linker into a single in-frame coding sequence (into a pSB1C3 backbone) using Gibson assembly. Success of the cloning procedure was confirmed by restriction with <i>EcoRI</i> and <i>SpeI</i> restriction enzymes followed by agarose gel electrophoresis (Figure 2) and in part by Sanger sequencing. This BioBrick was registered as part <a href="http://parts.igem.org/Part:BBa_K1846004">BBa_K1846004</a>. </p> | ||
| + | |||
| + | <img alt="File:BBK-stf-gene.jpg" src="/wiki/images/thumb/d/d7/BBK-stf-gene.jpg/515px-BBK-stf-gene.jpg.png" width="400" height="465" srcset="/wiki/images/thumb/d/d7/BBK-stf-gene.jpg/772px-BBK-stf-gene.jpg.png 1.5x, /wiki/images/thumb/d/d7/BBK-stf-gene.jpg/1029px-BBK-stf-gene.jpg.png 2x"> | ||
| + | |||
| + | <br><br><br> | ||
| + | |||
<br> | <br> | ||
| Line 24: | Line 30: | ||
<h3><b>tfa gene (BBa_K1846002)</b></h3> | <h3><b>tfa gene (BBa_K1846002)</b></h3> | ||
<img src="https://static.igem.org/mediawiki/2015/1/19/Birkbeck_tfa_phage.png"> | <img src="https://static.igem.org/mediawiki/2015/1/19/Birkbeck_tfa_phage.png"> | ||
| − | <p> | + | <p>The tfa (tail fibre assembly) protein of bacteriophage lambda assists in the assembly of the stf (short tail fibre) protein into a functional short tail fibre [4],[5]. Tfa gene displays a high level of homology (~40%) with the gp38 of bacteriophage T4 [1],[4],[6].</p> |
| + | <p>Sequence was optimised to remove illegal restriction sites. The gene sequence is currently available only as part of a composite part <a href="http://parts.igem.org/Part:BBa_K1846001">BBa_K1846001</a>.</p> | ||
<br> | <br> | ||
| Line 30: | Line 37: | ||
<h3><b>J gene (BBa_K1846008)</b></h3> | <h3><b>J gene (BBa_K1846008)</b></h3> | ||
<img src="https://static.igem.org/mediawiki/2015/b/b0/Birkbeck_Jgene_phage.png"> | <img src="https://static.igem.org/mediawiki/2015/b/b0/Birkbeck_Jgene_phage.png"> | ||
| − | <p>The J gene | + | <p>The bacteriophage lambda J gene encodes the tip attachment protein that forms the base plate of the bacteriophage's tail. It is responsible for host protein recognition and specificity. C-terminal residues of protein J have direct binding interactions with the maltoporin (LamB), a membrane protein of <i>E.coli</i>, allowing irreversible attachment of the phage to the host and facilitating DNA injection [3],[7]. |
| + | <p>We obtained this sequence as part of a collection of bacteriophage lambda parts from Technion, Haifa, Israel; this specific sequence was part of their previous BioBrick <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K784017">BBa_K784017</a>. The sequence contains a number of illegal PstI restriction sites. The BioBrick has been registered as part <a href="http://parts.igem.org/Part:BBa_K1846008">BBa_K1846008</a>, however we haven't been able to complete the removal of the restriction sites in time, so the part has not as yet been submitted to the registry.</p> | ||
<br> | <br> | ||
<br> | <br> | ||
| − | |||
| − | |||
| + | <b>References</b> | ||
| + | <p>[1] Hendrix, R. W., & Duda, R. L. (1992). Bacteriophage lambda PaPa: not the mother of all lambda phages. Science (New York, N.Y.), 258(5085), 1145–1148. doi:10.1126/science.1439823</p> | ||
| + | <p>[2] Haggard-Ljungquist, E., Halling, C., & Calendar, R. (1992). DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. Journal of Bacteriology, 174(5), 1462–1477.</p> | ||
| + | <p>[3] Chatterjee, S., & Rothenberg, E. (2012). Interaction of bacteriophage λ with its E. coli receptor, LamB. Viruses, 4(11), 3162–3178. doi:10.3390/v4113162</p> | ||
| + | <p>[4] Montag, D., Schwarz, H., & Henning, U. (1989). A component of the side tail fiber of E. coli bacteriophage λ can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. Journal of Bacteriology, 171(8), 4378–4384.</p> | ||
| + | <p>[5] Hashemolhosseini, S., Stierhof, Y. D., Hindennach, I., & Henning, U. (1996). Characterization of the helper proteins for the assembly of tail fibers of coliphages T4 and λ. Journal of Bacteriology, 178(21), 6258–6265.</p> | ||
| + | <p>[6] Haggard-Ljungquist, E., Halling, C., & Calendar, R. (1992). DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. Journal of Bacteriology, 174(5), 1462–1477.</p> | ||
| + | <p>[7] Werts, C., Michel, V., Hofnung, M., & Charbit, A. (1994). Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. Journal of Bacteriology, 176(4), 941–947.</p> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <li><a href="https://2015.igem.org/Team:Birkbeck/BioBricks">Return to BioBricks overview</a></li> | ||
| + | <li><a href="https://2015.igem.org/Team:Birkbeck/Composite_Part">Have a look at our Composite Parts</a></li> | ||
| + | </br></br> | ||
</div> | </div> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 21:29, 16 November 2015
Our BioBricks
Basic Parts
ORF314 (BBa_K1846000)

This is host-specific subunit of the short tail fibre protein of bacteriophage lambda. The C-terminus of the protein (ORF-314) contains the recognition site that confers binding specificity to the OmpC membrane protein of E.coli[1].
To transform ORF-314 sequence into a BioBrick basic part, we had the coding sequence synthesised, including the BioBrick prefix and suffix. The sequence was optimised to remove illegal restriction sites. We restricted both our synthesised ORF-314 and the linearised plasmid backbones (pSB1C3 for shipping, and pSB1K3 for further processing) with EcoRI and PstI restriction enzymes, before ligating the two pieces together using T4 DNA ligase. Success of the cloning procedure was confirmed by restriction with EcoRI and PstI restriction enzymes followed by agarose gel electrophoresis (Figure 1), with pSB1C3 alone and ORF314 alone as controls. The construct was also confirmed by sequencing. ORF-314 was submitted as BBa_K1846000.

Figure 1. Agarose gel electrophoresis of ORF314 inserted in pSB1C3 and restricted with EcoRI and PstI. The expected band sizes are 1 kb for ORF314 and 2 kb for pSB1C3. Successful results were obtained for samples 2 and 3.
stf (short tail fibre) gene (BBa_K1846004)

The stf gene encodes the short tail fibres found in the Ur-lambda bacteriophage, the original isolate [1]. The wild type lambda strain (λ-PaPa) that is most commonly used in research carries a frameshift mutation in the stf gene sequence producing no such fibres. It has been shown that the presence of these additional thin tails results in the more efficient adsorption to the host, E.coli [1].
The stf gene is a product of two open reading frames: ORF-401 and ORF-314. Introduction of 1 bp just after ORF-401 removes the frameshift fusing the two coding regions together which results in a functional short tail fibre protein consisting of 774 aa [2]. The C-terminal end of the stf protein (i.e. ORF-314) offers host receptor recognition for OmpC (Outer membrane protein C) on the surface of E.coli, widening receptor specificity and host range relative to λ-PaPa which only recognises the E.coli Maltoporin (lamB gene product) through its tip attachment protein J [1],[3]. Additionally, the C-terminus (ORF-314) displays a high level of homology with the gp37 of bacteriophage T4 [4].
The gene has been assembled using the synthesised ORF-401 with a linker sequence (removing the frameshift mutation) and ORF-314. The sequences were optimised to remove illegal restriction sites to make the sequence BioBrick-compatible. We cloned the two open reading frames and a linker into a single in-frame coding sequence (into a pSB1C3 backbone) using Gibson assembly. Success of the cloning procedure was confirmed by restriction with EcoRI and SpeI restriction enzymes followed by agarose gel electrophoresis (Figure 2) and in part by Sanger sequencing. This BioBrick was registered as part BBa_K1846004.
tfa gene (BBa_K1846002)

The tfa (tail fibre assembly) protein of bacteriophage lambda assists in the assembly of the stf (short tail fibre) protein into a functional short tail fibre [4],[5]. Tfa gene displays a high level of homology (~40%) with the gp38 of bacteriophage T4 [1],[4],[6].
Sequence was optimised to remove illegal restriction sites. The gene sequence is currently available only as part of a composite part BBa_K1846001.
J gene (BBa_K1846008)

The bacteriophage lambda J gene encodes the tip attachment protein that forms the base plate of the bacteriophage's tail. It is responsible for host protein recognition and specificity. C-terminal residues of protein J have direct binding interactions with the maltoporin (LamB), a membrane protein of E.coli, allowing irreversible attachment of the phage to the host and facilitating DNA injection [3],[7].
We obtained this sequence as part of a collection of bacteriophage lambda parts from Technion, Haifa, Israel; this specific sequence was part of their previous BioBrick BBa_K784017. The sequence contains a number of illegal PstI restriction sites. The BioBrick has been registered as part BBa_K1846008, however we haven't been able to complete the removal of the restriction sites in time, so the part has not as yet been submitted to the registry.
References
[1] Hendrix, R. W., & Duda, R. L. (1992). Bacteriophage lambda PaPa: not the mother of all lambda phages. Science (New York, N.Y.), 258(5085), 1145–1148. doi:10.1126/science.1439823
[2] Haggard-Ljungquist, E., Halling, C., & Calendar, R. (1992). DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. Journal of Bacteriology, 174(5), 1462–1477.
[3] Chatterjee, S., & Rothenberg, E. (2012). Interaction of bacteriophage λ with its E. coli receptor, LamB. Viruses, 4(11), 3162–3178. doi:10.3390/v4113162
[4] Montag, D., Schwarz, H., & Henning, U. (1989). A component of the side tail fiber of E. coli bacteriophage λ can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. Journal of Bacteriology, 171(8), 4378–4384.
[5] Hashemolhosseini, S., Stierhof, Y. D., Hindennach, I., & Henning, U. (1996). Characterization of the helper proteins for the assembly of tail fibers of coliphages T4 and λ. Journal of Bacteriology, 178(21), 6258–6265.
[6] Haggard-Ljungquist, E., Halling, C., & Calendar, R. (1992). DNA sequences of the tail fiber genes of bacteriophage P2: Evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. Journal of Bacteriology, 174(5), 1462–1477.
[7] Werts, C., Michel, V., Hofnung, M., & Charbit, A. (1994). Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. Journal of Bacteriology, 176(4), 941–947.




