Difference between revisions of "Team:Toronto/Notebook"
| (14 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | <!-- ####################################################### --> | ||
| + | <!-- # This html was produced by the igemwiki generator # --> | ||
| + | <!-- # https://github.com/igemuoftATG/generator-igemwiki # --> | ||
| + | <!-- ####################################################### --> | ||
| + | |||
| + | <!-- repo for this wiki: https://github.com/igemuoftATG/wiki2015 --> | ||
| + | |||
| + | </html> | ||
{{Toronto/head}} | {{Toronto/head}} | ||
<html> | <html> | ||
| Line 4: | Line 13: | ||
<div id="navigation"> | <div id="navigation"> | ||
<ul> | <ul> | ||
| − | <li><a href="https://2015.igem.org/Team:Toronto">home</a></li> | + | <li><a href="https://2015.igem.org/Team:Toronto"><span>home</span><i class="fa fa-home"></i></a></li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/Team">team</a></li> | + | </li> |
| − | <li><a href="#">project</a> | + | <li><a href="https://2015.igem.org/Team:Toronto/Team"><span>team</span><i class="fa fa-users"></i></a></li> |
| + | </li> | ||
| + | <li><a href="#"><span>project</span><i class="fa fa-info-circle"></i></a> | ||
<ul> | <ul> | ||
| − | <li><a href="https://2015.igem.org/Team:Toronto/Description">description</a></li> | + | <li><a href="https://2015.igem.org/Team:Toronto/Description"><span>description</span></a></li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/Experiments">experiments</a></li> | + | </li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/Results">results</a></li> | + | <li><a href="https://2015.igem.org/Team:Toronto/Experiments"><span>experiments</span></a></li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/Design">design</a></li> | + | </li> |
| + | <li><a href="https://2015.igem.org/Team:Toronto/Results"><span>results</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Design"><span>design</span></a></li> | ||
| + | </li> | ||
</ul> | </ul> | ||
| − | </li><li><a href="#"> | + | <li><a href="https://2015.igem.org/Team:Toronto/Parts"><span>parts</span><i class="fa fa-gear"></i></a></li> |
| + | </li> | ||
| + | <li class="active"><a href="https://2015.igem.org/Team:Toronto/Notebook"><span>notebook</span><i class="fa fa-book"></i></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Attributions"><span>attributions</span><i class="fa fa-life-saver"></i></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Collaborations"><span>collaborations</span><i class="fa fa-users"></i></a></li> | ||
| + | </li> | ||
| + | <li><a href="#"><span>practices</span><i class="fa fa-tree"></i></a> | ||
<ul> | <ul> | ||
| − | <li><a href="https://2015.igem.org/Team:Toronto/ | + | <li><a href="https://2015.igem.org/Team:Toronto/Practices"><span>human practices</span></a></li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/ | + | </li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/ | + | <li><a href="https://2015.igem.org/Team:Toronto/Alberta-Oil-Stories"><span>alberta oil stories</span></a></li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/ | + | </li> |
| + | <li><a href="https://2015.igem.org/Team:Toronto/Community-Outreach"><span>community outreach</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Technology-Applications"><span>technology applications</span></a></li> | ||
| + | </li> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/TechnoMoral-Scenarios"><span>technoMoral scenarios</span></a></li> | ||
| + | </li> | ||
</ul> | </ul> | ||
| − | < | + | <li><a href="https://2015.igem.org/Team:Toronto/Safety"><span>safety</span><i class="fa fa-fire"></i></a></li> |
| − | < | + | </li> |
| − | <li><a href=" | + | <li><a href="#"><span>software</span><i class="fa fa-code"></i></a> |
| − | < | + | <ul> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/ | + | <li><a href="https://2015.igem.org/Team:Toronto/Software"><span>software</span></a></li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/ | + | </li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/ | + | <li><a href="https://2015.igem.org/Team:Toronto/Demo"><span>demo</span></a></li> |
| − | < | + | </li> |
| − | <li><a href="https://2015.igem.org/Team:Toronto/Entrepreneurship">entrepreneurship</a></li> | + | <li><a href="https://2015.igem.org/Team:Toronto/Generator"><span>wiki generator</span></a></li> |
| + | </li> | ||
| + | </ul> | ||
| + | <li><a href="https://2015.igem.org/Team:Toronto/Entrepreneurship"><span>entrepreneurship</span><i class="fa fa-institution"></i></a></li> | ||
| + | </li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
| − | <div class="content"> | + | <div class="content" id="content-main"><div class="row"><div class="col col-lg-8 col-md-12"><div class="content-main"><h1 id="notebook">Notebook</h1> |
| − | < | + | <p>"The prefect lab notebook is a noble goal that very few achieve. But we must |
| − | <p> | + | all do our best."</p> |
| − | < | + | <h2 id="june">June</h2> |
| + | <h3 id="week-one">Week one</h3> | ||
| + | <h4 id="15-06-15">15/06/15</h4> | ||
<ul> | <ul> | ||
| − | <li> | + | <li>Prepared 8 x 500mL bottles of LB media</li> |
| − | <li> | + | <li>34 x 1mL Chloramphenicol (CAM) stocks</li> |
| − | <li> | + | <li>500mL of SOC media</li> |
| − | <li> | + | <li>Made LB plates</li> |
| + | <li>Made 70% EtOH</li> | ||
</ul> | </ul> | ||
| − | <h4 id=" | + | <h4 id="16-06-15">16/06/15</h4> |
| − | + | ||
<ul> | <ul> | ||
| − | <li> | + | <li>Made SOC media</li> |
| − | <li> | + | <li>Obtained DH10 β cells</li> |
| − | + | ||
| − | + | ||
</ul> | </ul> | ||
| − | + | <h4 id="17-06-15">17/06/15</h4> | |
| − | </div> | + | <ul> |
| + | <li>Bacterial transformation of RFP to test CAM stocks</li> | ||
| + | <li>Mark made more LB and Agar</li> | ||
| + | <li>Made overnight cultures of DH10 β</li> | ||
| + | </ul> | ||
| + | <h4 id="18-06-15">18/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Prepared GYT for making electrocompetent cells</li> | ||
| + | <li>Prepared LB and Agar</li> | ||
| + | <li>Observed growth of plated RFP</li> | ||
| + | <li>Made overnight cultures of RFP from plates</li> | ||
| + | </ul> | ||
| + | <h4 id="19-06-15">19/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Prepared 40% glycerol for glycerol stocks</li> | ||
| + | <li>Made glycerol stocks of MG1655 and RFP with pSB1C3 backbone</li> | ||
| + | <li>Made chemically competent cells.</li> | ||
| + | </ul> | ||
| + | <h3 id="week-two">Week Two</h3> | ||
| + | <h4 id="22-06-15">22/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Prepared TSS Buffer for making chemically competent cells</li> | ||
| + | <li>Made King's B media plates</li> | ||
| + | <li>Prepared electrocompetent DH10 β</li> | ||
| + | <li>Overnight cultures of transformed DH10β</li> | ||
| + | <li>Made LB plates with CAM</li> | ||
| + | </ul> | ||
| + | <h4 id="23-06-15">23/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Miniprep of cultured bacteria</li> | ||
| + | <li>GLycerol stocks of RFP plasmid</li> | ||
| + | <li>Casting of the gel for Gel Electrophoresis, as well as loading dye preparation</li> | ||
| + | <li>Made chemically competent cells, stored at -80C</li> | ||
| + | </ul> | ||
| + | <h4 id="24-06-15">24/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Made SOC media</li> | ||
| + | <li>Bacterial transformation</li> | ||
| + | <li>Preparation for gel electrophoresis.</li> | ||
| + | </ul> | ||
| + | <h4 id="25-06-15">25/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Prepared two overnight cultures of Pseudomonas putida F1 (PpF1) in King's B media</li> | ||
| + | <li>Plated PpF1 onto LB and 2 X King's B plates</li> | ||
| + | <li>Obtained:<ul> | ||
| + | <li>pDTG601 - with todC1C2BA insert</li> | ||
| + | <li>pDTG602 - with todC1C2BAD insert</li> | ||
| + | <li>pDTG603 - with todC1C2BADE insert</li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li>Stored obtained bacteria in the freezer</li> | ||
| + | </ul> | ||
| + | <h4 id="26-06-15">26/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Made 70% glycerol stocks of PpF1</li> | ||
| + | <li>Bacterial transformation with RFP</li> | ||
| + | </ul> | ||
| + | <h3 id="week-three">Week Three</h3> | ||
| + | <h4 id="29-06-15">29/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Made LB + Agar</li> | ||
| + | <li>Plated DH10 β</li> | ||
| + | </ul> | ||
| + | <h4 id="30-06-15">30/06/15</h4> | ||
| + | <ul> | ||
| + | <li>Transportation of Dangerous Goods (TDG) training</li> | ||
| + | </ul> | ||
| + | <h4 id="01-07-15">01/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Canada Day - Holiday</li> | ||
| + | </ul> | ||
| + | <h4 id="02-07-15">02/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Restriction enzyme digest for Interlab study</li> | ||
| + | <li>Prepared LB + Amp plates</li> | ||
| + | </ul> | ||
| + | <h4 id="03-07-15">03/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel Electrophoresis</li> | ||
| + | </ul> | ||
| + | <h2 id="july">July</h2> | ||
| + | <h3 id="week-four">Week Four</h3> | ||
| + | <h4 id="06-07-15">06/07/15</h4> | ||
| + | <h4 id="07-07-15">07/07/15</h4> | ||
| + | <ul> | ||
| + | <li>gel electrophoresis of Device1 A and Device1</li> | ||
| + | <li>ligation of device 1</li> | ||
| + | </ul> | ||
| + | <h4 id="08-07-15">08/07/15</h4> | ||
| + | <ul> | ||
| + | <li>gel electrophoresis of interlab device 2</li> | ||
| + | </ul> | ||
| + | <h4 id="09-07-15">09/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Bacterial transformation with interlab device 2</li> | ||
| + | <li>Plated transformed bacteria</li> | ||
| + | <li>Prepared Ampicillin stocks</li> | ||
| + | <li>Prepared 500mL of LB + Ampicillin (AMP) and stored at -4C</li> | ||
| + | <li>Prepared stocks of 70% Glycerol</li> | ||
| + | </ul> | ||
| + | <h4 id="10-07-15">10/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Miniprep of RFP and pDTG601</li> | ||
| + | <li>Gel electrophoresis of pDTG601 and RFP</li> | ||
| + | </ul> | ||
| + | <h3 id="week-five">Week Five</h3> | ||
| + | <h4 id="13-07-15">13/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Resuspended todF insert forward in 311uL of nuclease free water to a final 100uM solution</li> | ||
| + | <li>Resuspended todE insert forward in 311uL of nuclease free water to a final 100uM solution</li> | ||
| + | <li>Resuspended todF insert reverse in 300uL of nuclease free water to a final 100uM solution</li> | ||
| + | <li>Resuspended todF insert forward in 297uL of nuclease free water to a final 100uM solution</li> | ||
| + | <li>Resuspended todE insert in 100uL of nuclease free water</li> | ||
| + | <li><p>Resuspended todF insert in 100uL of nuclease free water</p> | ||
| + | </li> | ||
| + | <li><p>PCR amplified todE insert and todF insert using FroggaBio 2X Taq FroggaMix</p> | ||
| + | </li> | ||
| + | <li>Ran PCR purification of TodE and TodF according to the PureLink PCR Purification protocol.</li> | ||
| + | <li>Gel Electrophoresis of TodE and TodF PCR products.</li> | ||
| + | </ul> | ||
| + | <h4 id="14-07-15">14/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Prepared 250mL of SOC media</li> | ||
| + | <li>Prepared 25 LB + CAM plates</li> | ||
| + | <li>Gel electrophoresis of TodE and TodF PCR products</li> | ||
| + | </ul> | ||
| + | <h4 id="15-07-15">15/07/15</h4> | ||
| + | <ul> | ||
| + | <li>PCR amplified todE insert using FroggaBio 2X Taq FroggaMix</li> | ||
| + | <li>PCR purification of TodE</li> | ||
| + | <li>ran gel electrophoresis of TodE</li> | ||
| + | <li>Matt got stuck in the freight elevator.</li> | ||
| + | </ul> | ||
| + | <h4 id="16-07-15">16/07/15</h4> | ||
| + | <h4 id="17-07-15">17/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Gradient PCR for todE using FroggaBio 2X Taq FroggaMix</li> | ||
| + | </ul> | ||
| + | <h3 id="week-six">Week Six</h3> | ||
| + | <h4 id="20-07-15">20/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel electrophoresis</li> | ||
| + | <li>Cast SDS-PAGE gels</li> | ||
| + | </ul> | ||
| + | <h4 id="21-07-15">21/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Ran SDS-PAGE on RFP lysate</li> | ||
| + | <li>Made 40% glycerol stocks</li> | ||
| + | <li><p>Made TSS Buffer</p> | ||
| + | <h4 id="22-07-15">22/07/15</h4> | ||
| + | </li> | ||
| + | <li><p>Made chemically competent E.coli DH10β</p> | ||
| + | </li> | ||
| + | </ul> | ||
| + | <h4 id="23-07-15">23/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed bacterial transformation to test old batch of chemically competent cells against the newly made ones</li> | ||
| + | <li>A - old chemically competent cells with RFP + AMP resistence</li> | ||
| + | <li>B - new chemically competent cells with RFP + AMP resistence</li> | ||
| + | <li>Control - new chemically competent cells with RFP + CAM resistence</li> | ||
| + | <li>Ran day 2 of the cell lysis protocol of RFP and pDTG601 and pDTG603</li> | ||
| + | </ul> | ||
| + | <h4 id="24-07-15">24/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed Synbiota protocol for RDP assembly of GFP and aCP.</li> | ||
| + | </ul> | ||
| + | <h3 id="week-seven">Week Seven</h3> | ||
| + | <h4 id="27-07-15">27/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Prepared ligase for the P&P summer camp</li> | ||
| + | <li>Prepared lysozyme stocks</li> | ||
| + | <li>Performed day 3 of the cell lysis protocol</li> | ||
| + | <li>Prepared day 1 of the cell lysis protocol</li> | ||
| + | </ul> | ||
| + | <h4 id="28-07-15">28/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Preparation for SDS-PAGE, and gel Casting</li> | ||
| + | <li>Performed day 2 of the cell lysis Protocol</li> | ||
| + | </ul> | ||
| + | <h4 id="29-07-15">29/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed day 3 of the cell lysis Protocol</li> | ||
| + | <li>PCR amplification of the pSBC13 backbone</li> | ||
| + | <li>PCR purification of amplified backbone</li> | ||
| + | <li>Gel electrophoresis of the pSBC13 backbone</li> | ||
| + | </ul> | ||
| + | <h4 id="30-07-15">30/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed the PCR Protocol for creation of RDP parts with TodE and TodF inserts, using NEB Phusion polymerase.</li> | ||
| + | </ul> | ||
| + | <h4 id="31-07-15">31/07/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed PCR purification the Synbiota PCR TodE and TodF</li> | ||
| + | <li>Gel electrophoresis of PCR purified Synbiota TodE and TodF</li> | ||
| + | </ul> | ||
| + | <h2 id="august">August</h2> | ||
| + | <h3 id="week-eight">Week Eight</h3> | ||
| + | <h4 id="03-08-15">03/08/15</h4> | ||
| + | <ul> | ||
| + | <li>PCR amplification of pSB1C3 TodF using NEB Phusion polymerase</li> | ||
| + | </ul> | ||
| + | <h4 id="04-08-15">04/08/15</h4> | ||
| + | <ul> | ||
| + | <li>PCR purification of TodF and pSBC13</li> | ||
| + | <li>Synbiota BsaI digestion of TodE</li> | ||
| + | <li>Gel electrophoresis of BsaI digested TodE, pSB1C3 backbone, todF</li> | ||
| + | </ul> | ||
| + | <h4 id="05-08-15">05/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Synbiota PCR amplification of TodF and PCR of pSB1C3 backbone using Phusion polymerase</li> | ||
| + | <li>PCR purification</li> | ||
| + | <li>Miniprep of RFP</li> | ||
| + | </ul> | ||
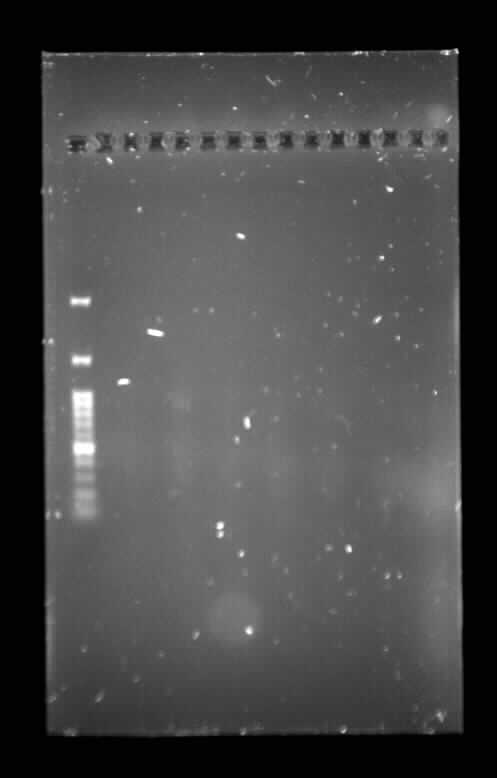
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_gel_todf_synbiota_aug_5.jpg]] <html> | ||
| + | </p> | ||
| + | TodF Gel - Synbiota August 5</p> | ||
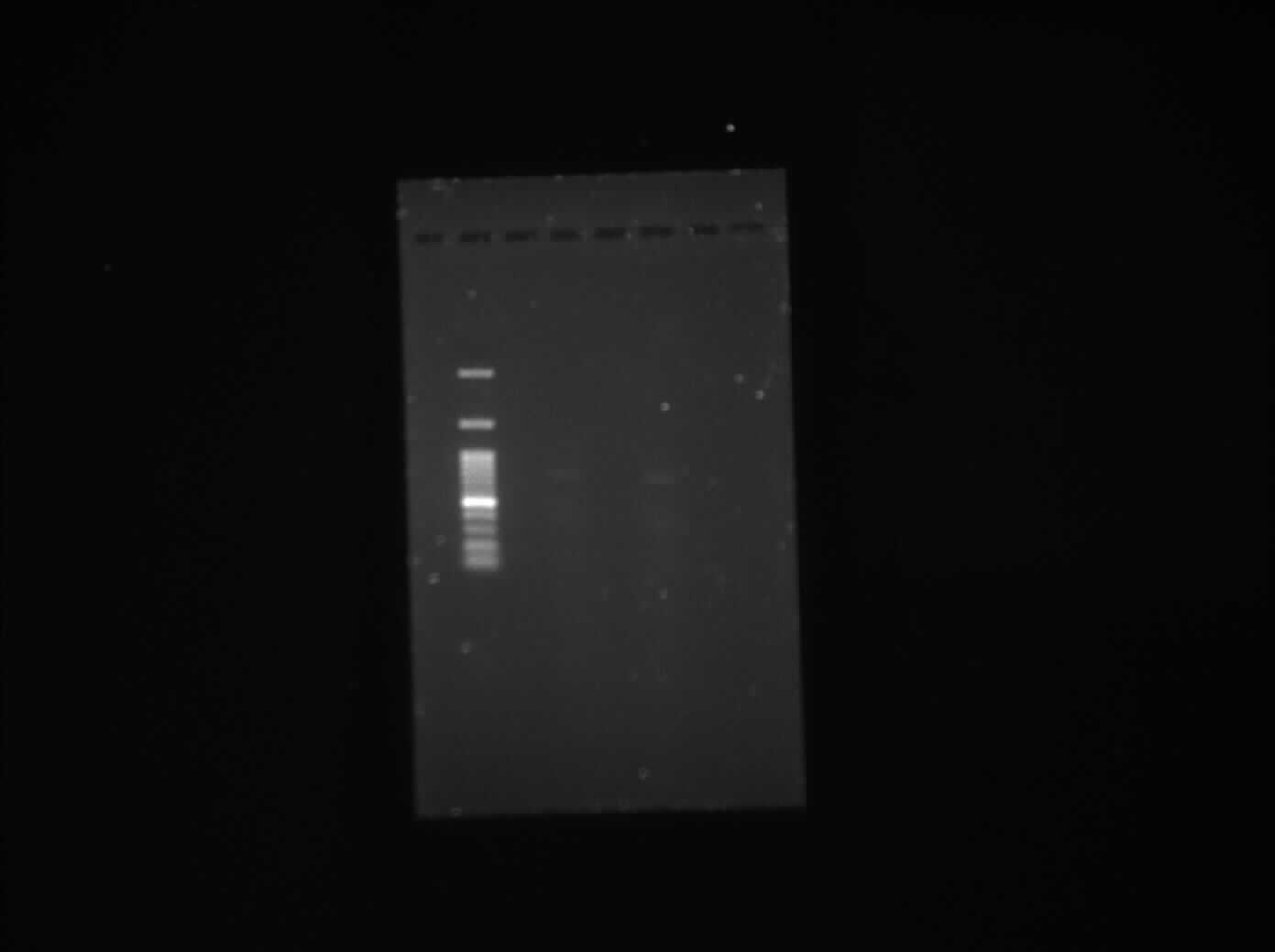
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_gel_tode_digested_synbiota_aug_5.jpg]] <html> | ||
| + | </p> | ||
| + | TodE Gel - Synbiota August 5</p> | ||
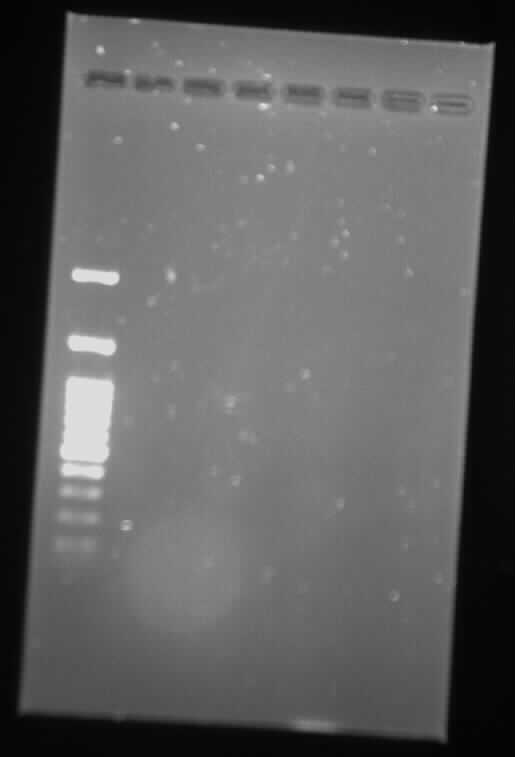
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_gel_synbiota_backbone_aug_5.jpg]] <html> | ||
| + | </p> | ||
| + | Gel Backbone - Synbiota August 5</p> | ||
| + | <h4 id="06-08-15">06/08/15</h4> | ||
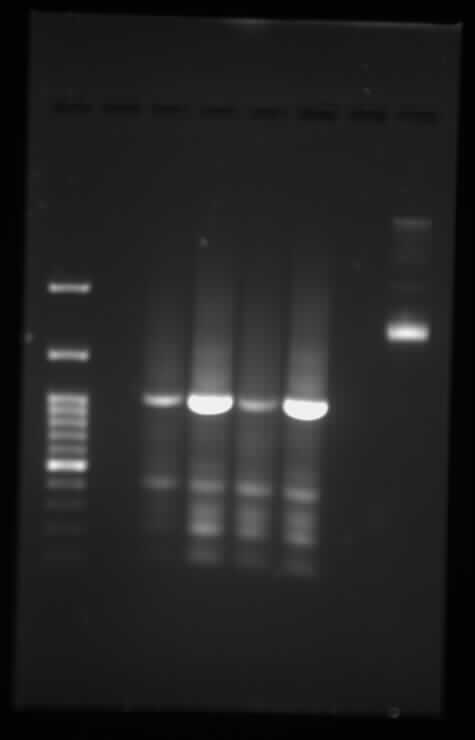
| + | <p>Gel electrophoresis of TodF, with IDT todF as a control</p> | ||
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_gel_todf_digested_synbiota_aug<em>6</em>(ladder-b-d-ladder-e-f-idtf).jpg]] <html> | ||
| + | </p> | ||
| + | TodF Gel Digested - Synbiota August 6</p> | ||
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_gel_todf_synbiota_aug<em>6</em>(ladder-b-d-ladder-e-f-idtf).jpg]] <html> | ||
| + | </p> | ||
| + | TodF Gel - Synbiota August 6</p> | ||
| + | <h4 id="07-08-15">07/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel electrophoresis of the pSB1C3 backbone</li> | ||
| + | <li>Synbiota RDP assembly of TodE and aCP</li> | ||
| + | <li>Re-tested competent cells with RFP</li> | ||
| + | </ul> | ||
| + | <h3 id="week-nine">Week Nine</h3> | ||
| + | <h4 id="10-08-15">10/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Ran SDS-PAGE with RFP, pDTG601, pDTG603 lysates</li> | ||
| + | <li>Observed results - high RFP expression</li> | ||
| + | <li>Preparation for Ligase Chain reaction (LCR)</li> | ||
| + | </ul> | ||
| + | <h4 id="11-08-15">11/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Miniprep of RFP</li> | ||
| + | <li>PCR amplification of pSB1C3 backbone with NEB Phusion polymerase.</li> | ||
| + | <li>PCR purification of the backbone</li> | ||
| + | <li>Made chemically competent cells</li> | ||
| + | </ul> | ||
| + | <h4 id="12-08-15">12/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Gradient PCR amplification of the pSB1C3 backbone using NEB Phusion polymerase and NEB Q5 High Fidelity polymerase (with and without GC enhancer).</li> | ||
| + | <li>Performed DPN1 Digest and PCR purifications of amplified pSB1C3 Protocol</li> | ||
| + | </ul> | ||
| + | <h4 id="13-08-15">13/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed bacterial transformations with miniprepped RFP and resuspended RFP (with both old and new competent cells, and old and new TSS buffer). All grew, miniprepped RFP and new TSS work best.</li> | ||
| + | <li>Gel electrophoresis of PCR pSB1C3 backbone with Q5 polymerase, and with Q5 polymerase and GC enhancer.</li> | ||
| + | <li>Performed LCR with TodE and TodF inserts</li> | ||
| + | <li>Performed PCR of TodF with Phusion polymerase.</li> | ||
| + | <li>Performed bacterial transformations with purified PCR products.</li> | ||
| + | </ul> | ||
| + | <h4 id="14-08-15">14/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Made 500mL of LB + Agar</li> | ||
| + | <li>made 70% EtOH</li> | ||
| + | <li>Made 1X TAE</li> | ||
| + | <li>PCR purification of amplified Synbiota TodF</li> | ||
| + | <li>Gel Electrophoresis of Synbiota TodF</li> | ||
| + | </ul> | ||
| + | <h3 id="week-ten">Week Ten</h3> | ||
| + | <h4 id="17-08-15">17/08/15</h4> | ||
| + | <ul> | ||
| + | <li>PCR of the pSBC13 backbone with new primers, using NEB Q5 polymerase</li> | ||
| + | <li>Gel electrophoresis of LCR TodE and TodF products</li> | ||
| + | <li>DPN1 digest and PCR purification of the pSB1C3 backbone</li> | ||
| + | <li>Gel electrophoresis of the pSB1C3 backbone</li> | ||
| + | </ul> | ||
| + | <h4 id="18-08-15">18/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed and LCR reaction with TodE and TodF inserts</li> | ||
| + | <li>Performed Synbiota assembly of TodE and aCP.</li> | ||
| + | <li>Performed bacterial transformation with LCR and Synbiota products, with RFP as a control.</li> | ||
| + | </ul> | ||
| + | <h4 id="19-08-15">19/08/15</h4> | ||
| + | <h4 id="20-08-15">20/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Synbiota assembly of GFP and aCP</li> | ||
| + | <li>Bacterial transformation with GFP, aCP and RFP as a control.</li> | ||
| + | </ul> | ||
| + | <h4 id="21-08-15">21/08/15</h4> | ||
| + | <h3 id="week-eleven">Week Eleven</h3> | ||
| + | <h4 id="24-08-15">24/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel electrophoresis of Synbiota aCP and GFP</li> | ||
| + | </ul> | ||
| + | <h4 id="25-08-15">25/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Performed an LCR reaction with TodE and TodF inserts.</li> | ||
| + | <li>Gibson assembly of TodE as well as with TodE and TodF insterts</li> | ||
| + | </ul> | ||
| + | <h4 id="26-08-15">26/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel electrophoresis of Gibson products of TodE and TodE & TodF</li> | ||
| + | </ul> | ||
| + | <h4 id="27-08-15">27/08/15</h4> | ||
| + | <h4 id="28-08-15">28/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel electrophoresis of Gibson products of TodE and TodE & TodF</li> | ||
| + | </ul> | ||
| + | <h2 id="september">September</h2> | ||
| + | <h3 id="week-twelve">Week Twelve</h3> | ||
| + | <h4 id="31-08-15">31/08/15</h4> | ||
| + | <ul> | ||
| + | <li>Bacterial transformation with LCR products, RFP as a control</li> | ||
| + | <li>PCR of Synbiota TodE with Phusion polymerase</li> | ||
| + | <li>Synbiota assembly of aCP and TodE</li> | ||
| + | </ul> | ||
| + | <h4 id="01-09-15">01/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Made 1xTAE</li> | ||
| + | <li>Bacterial transformation with LCR remnants, aCP, todE, RFP.</li> | ||
| + | <li>Gel electrophoresis of Synbiota TodE and aCP</li> | ||
| + | <li>Overnight cultures of LCR products, TodE, RFP</li> | ||
| + | </ul> | ||
| + | <h4 id="02-09-15">02/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Gibson assembly of TodE and TodF inserts.</li> | ||
| + | <li>1mL SOC aliquot stocks</li> | ||
| + | <li>Transformation with aCP, TodE, RFP</li> | ||
| + | </ul> | ||
| + | <h4 id="03-09-15">03/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Miniprep of LCR products, todE, RFP</li> | ||
| + | <li>Synbiota assembly of todE and aCP</li> | ||
| + | </ul> | ||
| + | <h4 id="04-09-15">04/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Synbiota assembly of TodE and aCP</li> | ||
| + | <li>Bacterial transformation with TodE from Synbiota and with TodE from Gibson assembly, Gibson assembled TodE and TodF</li> | ||
| + | </ul> | ||
| + | <h3 id="week-thirteen">Week Thirteen</h3> | ||
| + | <h4 id="07-09-15">07/09/15</h4> | ||
| + | <p>Labour Day</p> | ||
| + | <h4 id="08-09-15">08/09/15</h4> | ||
| + | <ul> | ||
| + | <li>gel electrophoresis for TodF of Gibson assembly</li> | ||
| + | </ul> | ||
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_TodF_gel_gibson_Sept-8.jpg]] <html> | ||
| + | </p> | ||
| + | TodF Gel - Gibson September 8th</p> | ||
| + | <h4 id="09-09-15">09/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel electrophoresis of TodF for Gibson assembly</li> | ||
| + | <li>Miniprep of RFP, TodE and aCP</li> | ||
| + | <li>Bacterial transformations with Gibson TodEF plasmids, Gibson todE plasmid, RFP and LCP products</li> | ||
| + | </ul> | ||
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_TodF_gel_gibson_TodE_Sept-9.jpg]] <html> | ||
| + | </p> | ||
| + | TodF Gel - Gibson TodE - September 9</p> | ||
| + | <h4 id="10-09-15">10/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Bacterial transformation with diluted Gibson products</li> | ||
| + | </ul> | ||
| + | <h4 id="11-09-15">11/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Synbiota assembly of TodE</li> | ||
| + | <li>Transformation with LCR product</li> | ||
| + | </ul> | ||
| + | <h3 id="week-fourteen">Week Fourteen</h3> | ||
| + | <h4 id="14-09-15">14/09/15</h4> | ||
| + | <ul> | ||
| + | <li>LCR of TodE and TodF</li> | ||
| + | </ul> | ||
| + | <p><p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_TodE_gel_Sept-14.jpg]] <html> | ||
| + | </p> | ||
| + | TodE Gel Sep. 14</p> | ||
| + | <h4 id="15-09-15">15/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Gel electrophoresis of LCR products</li> | ||
| + | </ul> | ||
| + | <h4 id="16-09-15">16/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Gibson assembly of TodE and TodF and of TodE</li> | ||
| + | <li>Bacterial transformation with LCR products and Giboson products</li> | ||
| + | </ul> | ||
| + | <h4 id="17-09-15">17/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Observed Growth of Gibson assembled TodE and prepared overnight cultures.</li> | ||
| + | </ul> | ||
| + | <h4 id="18-09-15">18/09/15</h4> | ||
| + | <ul> | ||
| + | <li>Miniprep of Gibson assembled todE plasmid</li> | ||
| + | <li>Preparing part for shipping to iGEM HQ, used a concentrator to speed vacuum our part.</li> | ||
| + | <li>Performed two restriction enzyme digests to verify for the presence of TodE</li> | ||
| + | <li>First digest with XhoI and XcmI produced appropriate band lengths.</li> | ||
| + | <li>The Second Digest was with RsaI and PvuII also confirmed TodE presence.</li> | ||
| + | </ul> | ||
| + | <!-- <p class="image-wrapper"> | ||
| + | </html> [[File:Toronto_2015_TodE_only_gel_Sept-18.jpg]] <html> | ||
| + | </p> --> | ||
| + | </div></div><div id="tableofcontents" class="tableofcontents affix sidebar col-lg-4 hidden-xs hidden-sm hidden-md visible-lg-3"><ul class="nav"> | ||
| + | <li><a href="#june">June</a><ul> | ||
| + | <li><a href="#week-one">Week one</a><ul> | ||
| + | <li><a href="#15-06-15">15/06/15</a></li> | ||
| + | <li><a href="#16-06-15">16/06/15</a></li> | ||
| + | <li><a href="#17-06-15">17/06/15</a></li> | ||
| + | <li><a href="#18-06-15">18/06/15</a></li> | ||
| + | <li><a href="#19-06-15">19/06/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-two">Week Two</a><ul> | ||
| + | <li><a href="#22-06-15">22/06/15</a></li> | ||
| + | <li><a href="#23-06-15">23/06/15</a></li> | ||
| + | <li><a href="#24-06-15">24/06/15</a></li> | ||
| + | <li><a href="#25-06-15">25/06/15</a></li> | ||
| + | <li><a href="#26-06-15">26/06/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-three">Week Three</a><ul> | ||
| + | <li><a href="#29-06-15">29/06/15</a></li> | ||
| + | <li><a href="#30-06-15">30/06/15</a></li> | ||
| + | <li><a href="#01-07-15">01/07/15</a></li> | ||
| + | <li><a href="#02-07-15">02/07/15</a></li> | ||
| + | <li><a href="#03-07-15">03/07/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#july">July</a><ul> | ||
| + | <li><a href="#week-four">Week Four</a><ul> | ||
| + | <li><a href="#06-07-15">06/07/15</a></li> | ||
| + | <li><a href="#07-07-15">07/07/15</a></li> | ||
| + | <li><a href="#08-07-15">08/07/15</a></li> | ||
| + | <li><a href="#09-07-15">09/07/15</a></li> | ||
| + | <li><a href="#10-07-15">10/07/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-five">Week Five</a><ul> | ||
| + | <li><a href="#13-07-15">13/07/15</a></li> | ||
| + | <li><a href="#14-07-15">14/07/15</a></li> | ||
| + | <li><a href="#15-07-15">15/07/15</a></li> | ||
| + | <li><a href="#16-07-15">16/07/15</a></li> | ||
| + | <li><a href="#17-07-15">17/07/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-six">Week Six</a><ul> | ||
| + | <li><a href="#20-07-15">20/07/15</a></li> | ||
| + | <li><a href="#21-07-15">21/07/15</a></li> | ||
| + | <li><a href="#22-07-15">22/07/15</a></li> | ||
| + | <li><a href="#23-07-15">23/07/15</a></li> | ||
| + | <li><a href="#24-07-15">24/07/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-seven">Week Seven</a><ul> | ||
| + | <li><a href="#27-07-15">27/07/15</a></li> | ||
| + | <li><a href="#28-07-15">28/07/15</a></li> | ||
| + | <li><a href="#29-07-15">29/07/15</a></li> | ||
| + | <li><a href="#30-07-15">30/07/15</a></li> | ||
| + | <li><a href="#31-07-15">31/07/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#august">August</a><ul> | ||
| + | <li><a href="#week-eight">Week Eight</a><ul> | ||
| + | <li><a href="#03-08-15">03/08/15</a></li> | ||
| + | <li><a href="#04-08-15">04/08/15</a></li> | ||
| + | <li><a href="#05-08-15">05/08/15</a></li> | ||
| + | <li><a href="#06-08-15">06/08/15</a></li> | ||
| + | <li><a href="#07-08-15">07/08/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-nine">Week Nine</a><ul> | ||
| + | <li><a href="#10-08-15">10/08/15</a></li> | ||
| + | <li><a href="#11-08-15">11/08/15</a></li> | ||
| + | <li><a href="#12-08-15">12/08/15</a></li> | ||
| + | <li><a href="#13-08-15">13/08/15</a></li> | ||
| + | <li><a href="#14-08-15">14/08/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-ten">Week Ten</a><ul> | ||
| + | <li><a href="#17-08-15">17/08/15</a></li> | ||
| + | <li><a href="#18-08-15">18/08/15</a></li> | ||
| + | <li><a href="#19-08-15">19/08/15</a></li> | ||
| + | <li><a href="#20-08-15">20/08/15</a></li> | ||
| + | <li><a href="#21-08-15">21/08/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-eleven">Week Eleven</a><ul> | ||
| + | <li><a href="#24-08-15">24/08/15</a></li> | ||
| + | <li><a href="#25-08-15">25/08/15</a></li> | ||
| + | <li><a href="#26-08-15">26/08/15</a></li> | ||
| + | <li><a href="#27-08-15">27/08/15</a></li> | ||
| + | <li><a href="#28-08-15">28/08/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#september">September</a><ul> | ||
| + | <li><a href="#week-twelve">Week Twelve</a><ul> | ||
| + | <li><a href="#31-08-15">31/08/15</a></li> | ||
| + | <li><a href="#01-09-15">01/09/15</a></li> | ||
| + | <li><a href="#02-09-15">02/09/15</a></li> | ||
| + | <li><a href="#03-09-15">03/09/15</a></li> | ||
| + | <li><a href="#04-09-15">04/09/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-thirteen">Week Thirteen</a><ul> | ||
| + | <li><a href="#07-09-15">07/09/15</a></li> | ||
| + | <li><a href="#08-09-15">08/09/15</a></li> | ||
| + | <li><a href="#09-09-15">09/09/15</a></li> | ||
| + | <li><a href="#10-09-15">10/09/15</a></li> | ||
| + | <li><a href="#11-09-15">11/09/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#week-fourteen">Week Fourteen</a><ul> | ||
| + | <li><a href="#14-09-15">14/09/15</a></li> | ||
| + | <li><a href="#15-09-15">15/09/15</a></li> | ||
| + | <li><a href="#16-09-15">16/09/15</a></li> | ||
| + | <li><a href="#17-09-15">17/09/15</a></li> | ||
| + | <li><a href="#18-09-15">18/09/15</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </div></div></div> | ||
</html> | </html> | ||
{{Toronto/footer}} | {{Toronto/footer}} | ||
Latest revision as of 04:32, 20 November 2015
Notebook
"The prefect lab notebook is a noble goal that very few achieve. But we must all do our best."
June
Week one
15/06/15
- Prepared 8 x 500mL bottles of LB media
- 34 x 1mL Chloramphenicol (CAM) stocks
- 500mL of SOC media
- Made LB plates
- Made 70% EtOH
16/06/15
- Made SOC media
- Obtained DH10 β cells
17/06/15
- Bacterial transformation of RFP to test CAM stocks
- Mark made more LB and Agar
- Made overnight cultures of DH10 β
18/06/15
- Prepared GYT for making electrocompetent cells
- Prepared LB and Agar
- Observed growth of plated RFP
- Made overnight cultures of RFP from plates
19/06/15
- Prepared 40% glycerol for glycerol stocks
- Made glycerol stocks of MG1655 and RFP with pSB1C3 backbone
- Made chemically competent cells.
Week Two
22/06/15
- Prepared TSS Buffer for making chemically competent cells
- Made King's B media plates
- Prepared electrocompetent DH10 β
- Overnight cultures of transformed DH10β
- Made LB plates with CAM
23/06/15
- Miniprep of cultured bacteria
- GLycerol stocks of RFP plasmid
- Casting of the gel for Gel Electrophoresis, as well as loading dye preparation
- Made chemically competent cells, stored at -80C
24/06/15
- Made SOC media
- Bacterial transformation
- Preparation for gel electrophoresis.
25/06/15
- Prepared two overnight cultures of Pseudomonas putida F1 (PpF1) in King's B media
- Plated PpF1 onto LB and 2 X King's B plates
- Obtained:
- pDTG601 - with todC1C2BA insert
- pDTG602 - with todC1C2BAD insert
- pDTG603 - with todC1C2BADE insert
- Stored obtained bacteria in the freezer
26/06/15
- Made 70% glycerol stocks of PpF1
- Bacterial transformation with RFP
Week Three
29/06/15
- Made LB + Agar
- Plated DH10 β
30/06/15
- Transportation of Dangerous Goods (TDG) training
01/07/15
- Canada Day - Holiday
02/07/15
- Restriction enzyme digest for Interlab study
- Prepared LB + Amp plates
03/07/15
- Gel Electrophoresis
July
Week Four
06/07/15
07/07/15
- gel electrophoresis of Device1 A and Device1
- ligation of device 1
08/07/15
- gel electrophoresis of interlab device 2
09/07/15
- Bacterial transformation with interlab device 2
- Plated transformed bacteria
- Prepared Ampicillin stocks
- Prepared 500mL of LB + Ampicillin (AMP) and stored at -4C
- Prepared stocks of 70% Glycerol
10/07/15
- Miniprep of RFP and pDTG601
- Gel electrophoresis of pDTG601 and RFP
Week Five
13/07/15
- Resuspended todF insert forward in 311uL of nuclease free water to a final 100uM solution
- Resuspended todE insert forward in 311uL of nuclease free water to a final 100uM solution
- Resuspended todF insert reverse in 300uL of nuclease free water to a final 100uM solution
- Resuspended todF insert forward in 297uL of nuclease free water to a final 100uM solution
- Resuspended todE insert in 100uL of nuclease free water
Resuspended todF insert in 100uL of nuclease free water
PCR amplified todE insert and todF insert using FroggaBio 2X Taq FroggaMix
- Ran PCR purification of TodE and TodF according to the PureLink PCR Purification protocol.
- Gel Electrophoresis of TodE and TodF PCR products.
14/07/15
- Prepared 250mL of SOC media
- Prepared 25 LB + CAM plates
- Gel electrophoresis of TodE and TodF PCR products
15/07/15
- PCR amplified todE insert using FroggaBio 2X Taq FroggaMix
- PCR purification of TodE
- ran gel electrophoresis of TodE
- Matt got stuck in the freight elevator.
16/07/15
17/07/15
- Gradient PCR for todE using FroggaBio 2X Taq FroggaMix
Week Six
20/07/15
- Gel electrophoresis
- Cast SDS-PAGE gels
21/07/15
- Ran SDS-PAGE on RFP lysate
- Made 40% glycerol stocks
Made TSS Buffer
22/07/15
Made chemically competent E.coli DH10β
23/07/15
- Performed bacterial transformation to test old batch of chemically competent cells against the newly made ones
- A - old chemically competent cells with RFP + AMP resistence
- B - new chemically competent cells with RFP + AMP resistence
- Control - new chemically competent cells with RFP + CAM resistence
- Ran day 2 of the cell lysis protocol of RFP and pDTG601 and pDTG603
24/07/15
- Performed Synbiota protocol for RDP assembly of GFP and aCP.
Week Seven
27/07/15
- Prepared ligase for the P&P summer camp
- Prepared lysozyme stocks
- Performed day 3 of the cell lysis protocol
- Prepared day 1 of the cell lysis protocol
28/07/15
- Preparation for SDS-PAGE, and gel Casting
- Performed day 2 of the cell lysis Protocol
29/07/15
- Performed day 3 of the cell lysis Protocol
- PCR amplification of the pSBC13 backbone
- PCR purification of amplified backbone
- Gel electrophoresis of the pSBC13 backbone
30/07/15
- Performed the PCR Protocol for creation of RDP parts with TodE and TodF inserts, using NEB Phusion polymerase.
31/07/15
- Performed PCR purification the Synbiota PCR TodE and TodF
- Gel electrophoresis of PCR purified Synbiota TodE and TodF
August
Week Eight
03/08/15
- PCR amplification of pSB1C3 TodF using NEB Phusion polymerase
04/08/15
- PCR purification of TodF and pSBC13
- Synbiota BsaI digestion of TodE
- Gel electrophoresis of BsaI digested TodE, pSB1C3 backbone, todF
05/08/15
- Synbiota PCR amplification of TodF and PCR of pSB1C3 backbone using Phusion polymerase
- PCR purification
- Miniprep of RFP
06/08/15
Gel electrophoresis of TodF, with IDT todF as a control
[[File:Toronto_2015_gel_todf_digested_synbiota_aug6(ladder-b-d-ladder-e-f-idtf).jpg]]
TodF Gel Digested - Synbiota August 6[[File:Toronto_2015_gel_todf_synbiota_aug6(ladder-b-d-ladder-e-f-idtf).jpg]]
TodF Gel - Synbiota August 607/08/15
- Gel electrophoresis of the pSB1C3 backbone
- Synbiota RDP assembly of TodE and aCP
- Re-tested competent cells with RFP
Week Nine
10/08/15
- Ran SDS-PAGE with RFP, pDTG601, pDTG603 lysates
- Observed results - high RFP expression
- Preparation for Ligase Chain reaction (LCR)
11/08/15
- Miniprep of RFP
- PCR amplification of pSB1C3 backbone with NEB Phusion polymerase.
- PCR purification of the backbone
- Made chemically competent cells
12/08/15
- Gradient PCR amplification of the pSB1C3 backbone using NEB Phusion polymerase and NEB Q5 High Fidelity polymerase (with and without GC enhancer).
- Performed DPN1 Digest and PCR purifications of amplified pSB1C3 Protocol
13/08/15
- Performed bacterial transformations with miniprepped RFP and resuspended RFP (with both old and new competent cells, and old and new TSS buffer). All grew, miniprepped RFP and new TSS work best.
- Gel electrophoresis of PCR pSB1C3 backbone with Q5 polymerase, and with Q5 polymerase and GC enhancer.
- Performed LCR with TodE and TodF inserts
- Performed PCR of TodF with Phusion polymerase.
- Performed bacterial transformations with purified PCR products.
14/08/15
- Made 500mL of LB + Agar
- made 70% EtOH
- Made 1X TAE
- PCR purification of amplified Synbiota TodF
- Gel Electrophoresis of Synbiota TodF
Week Ten
17/08/15
- PCR of the pSBC13 backbone with new primers, using NEB Q5 polymerase
- Gel electrophoresis of LCR TodE and TodF products
- DPN1 digest and PCR purification of the pSB1C3 backbone
- Gel electrophoresis of the pSB1C3 backbone
18/08/15
- Performed and LCR reaction with TodE and TodF inserts
- Performed Synbiota assembly of TodE and aCP.
- Performed bacterial transformation with LCR and Synbiota products, with RFP as a control.
19/08/15
20/08/15
- Synbiota assembly of GFP and aCP
- Bacterial transformation with GFP, aCP and RFP as a control.
21/08/15
Week Eleven
24/08/15
- Gel electrophoresis of Synbiota aCP and GFP
25/08/15
- Performed an LCR reaction with TodE and TodF inserts.
- Gibson assembly of TodE as well as with TodE and TodF insterts
26/08/15
- Gel electrophoresis of Gibson products of TodE and TodE & TodF
27/08/15
28/08/15
- Gel electrophoresis of Gibson products of TodE and TodE & TodF
September
Week Twelve
31/08/15
- Bacterial transformation with LCR products, RFP as a control
- PCR of Synbiota TodE with Phusion polymerase
- Synbiota assembly of aCP and TodE
01/09/15
- Made 1xTAE
- Bacterial transformation with LCR remnants, aCP, todE, RFP.
- Gel electrophoresis of Synbiota TodE and aCP
- Overnight cultures of LCR products, TodE, RFP
02/09/15
- Gibson assembly of TodE and TodF inserts.
- 1mL SOC aliquot stocks
- Transformation with aCP, TodE, RFP
03/09/15
- Miniprep of LCR products, todE, RFP
- Synbiota assembly of todE and aCP
04/09/15
- Synbiota assembly of TodE and aCP
- Bacterial transformation with TodE from Synbiota and with TodE from Gibson assembly, Gibson assembled TodE and TodF
Week Thirteen
07/09/15
Labour Day
08/09/15
- gel electrophoresis for TodF of Gibson assembly
09/09/15
- Gel electrophoresis of TodF for Gibson assembly
- Miniprep of RFP, TodE and aCP
- Bacterial transformations with Gibson TodEF plasmids, Gibson todE plasmid, RFP and LCP products
10/09/15
- Bacterial transformation with diluted Gibson products
11/09/15
- Synbiota assembly of TodE
- Transformation with LCR product
Week Fourteen
14/09/15
- LCR of TodE and TodF
15/09/15
- Gel electrophoresis of LCR products
16/09/15
- Gibson assembly of TodE and TodF and of TodE
- Bacterial transformation with LCR products and Giboson products
17/09/15
- Observed Growth of Gibson assembled TodE and prepared overnight cultures.
18/09/15
- Miniprep of Gibson assembled todE plasmid
- Preparing part for shipping to iGEM HQ, used a concentrator to speed vacuum our part.
- Performed two restriction enzyme digests to verify for the presence of TodE
- First digest with XhoI and XcmI produced appropriate band lengths.
- The Second Digest was with RsaI and PvuII also confirmed TodE presence.