Difference between revisions of "Team:Waterloo/Lab/dCas9"
(add visualization) |
|||
| (11 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
<h1>dCas9 Modification</h1> | <h1>dCas9 Modification</h1> | ||
| + | <section id="intro" title="Introduction"> | ||
| + | <h2>Introduction</h2> | ||
| + | <p> | ||
| + | One of our goals this year was to engineer a version of the Cas9 protein that could recognize different protospacer adjacent motifs (PAMs) other than the regular NGG. Kleinstiver <em>et al.</em> <cite ref="Kleinstiver2015"></cite> recently published data on Cas9 mutants that effectively cut unique PAM sites. A specific EQR variant in the publication recognized a NGAG PAM site after three different amino acids in the original Cas9 were changed. The EQR variant includes the changes D1135E, R1335Q, and T1337R. Our goal was to try and replicate these mutations but instead with dCas9, which is commonly used since it represses a gene rather than cutting one. Target sites were designed in promoters of green fluorescent protein (GFP) with both an NGG PAM sequence and an NGAG PAM sequence. | ||
| + | </p> | ||
| + | </section> | ||
| − | < | + | <section id="design" title="Experimental Design"> |
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | <p> | + | <h2> Experimental Design </h2> |
| − | Our first step was to create the EQR variant in dCas9 by using site directed mutagenesis | + | <p> |
| − | </p> | + | All of our experiments to study GFP expression with and without addition of dCas9 were performed on the Amnis Image Stream machine (Amnis 2015). The Image Stream is a fluorescence microscope combined with a flow cytometer, letting one to perform a quantitative method of fluorescence detection while visualizing the actual cells. By measuring an average fluorescence intensity of cells, we can determine the average intensity of a cell population. |
| + | </p> | ||
| + | <p> | ||
| + | Our first step was to create the EQR variant in dCas9 by using <a href="https://2015.igem.org/Team:Waterloo/Experiments">site directed mutagenesis</a>. This was done on a dCas9 plasmid ordered from AddGene. Two other plasmids were constructed with both a gene producing GFP and an sgRNA target recognizing either an NGG or an NGAG PAM site on the GFP promoter J23101. | ||
| + | </p> | ||
| + | <p> | ||
| + | The first experiment that was performed was to co-transform a plasmid containing both GFP and the sgRNA targeting the GFP promoter into the same cell as dCas9. This was a control experiment that will explain that the mutated EQR dCas9 performs worse than | ||
| + | the original dCas9 when targeting an NGG PAM site. | ||
| + | </p> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/5/54/Waterloo_ImageNGG.png"> | ||
| + | </figure> | ||
| + | <p> | ||
| + | The next experiment involved transforming the plasmid with GFP and an sgRNA targeting a region in the GFP promoter with an NGAG PAM site into <em>E. coli</em>. This was also co-transformed with both the orignial and the mutated versions of dCas9. This time, we expect to see the mutated EQR dCas9 perform better than the original in terms of repression of GFP. | ||
| + | </p> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/d/dc/Waterloo_ImageNGAG.png"> | ||
| + | </figure> | ||
| + | </section> | ||
| + | <section id="constructs" title="Constructs"> | ||
| + | <h2> Constructs </h2> | ||
| − | < | + | <figure> |
| + | <img src="/wiki/images/e/ec/Waterloo_pamflexlabchange.png" alt="Visualization of site-directed mutagenesis" /> | ||
| + | <figcaption>Visualization of site-directed mutagenesis protocol, showing modification to dCas9 coding sequence.</figcaption> | ||
| + | </figure> | ||
| − | < | + | <ul> |
| − | Target | + | <li>GFP with NGAG sgRNA target</li> |
| − | Mutated dCas9 | + | <li>Target plasmid with GFP sgRNA</li> |
| − | + | <li>Mutated dCas9</li> | |
| − | < | + | <li>dCas9 from AddGene</li> |
| + | </ul> | ||
| + | </section> | ||
| − | <h2> Results </h2> | + | <section id="results" title="Results"> |
| + | <h2> Results </h2> | ||
| + | <p> | ||
| + | The EQR muatated dCas9 was successfully constructed using the QuickChange protocol twice and sending for sequencing. Co-transformations were also accomplished and cells were analyzed using Flow cytometry. | ||
| + | </p> | ||
| + | <h3> NGG PAM site </h3> | ||
| + | <div class="jumbotron hp-landing"> | ||
| + | <div class="container"> | ||
| + | <div class="row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/e/e6/Waterloo_flow_orig_NGG.png"> | ||
| + | </figure> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/1/16/Waterloo_flow_mut_NGG.png"> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <h3> NGAG PAM site </h3> | ||
| + | <div class="jumbotron hp-landing"> | ||
| + | <div class="container"> | ||
| + | <div class="row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/8/87/Waterloo_flow_mut_NGaG.png"> | ||
| + | </figure> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/1/1b/Waterloo_flow_orig_NGaG.png"> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <p> | ||
| + | These graphs show the results of the mean of the GFP intensity when the co-transformed cells were run on flow cytometry. A lower mean points to the result that the dCas9 is more effective at repressing expression of GFP. These results are inconclusive and we need to repeat the experiment multiple times to understand the effect of the EQR mutations on dCas9. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
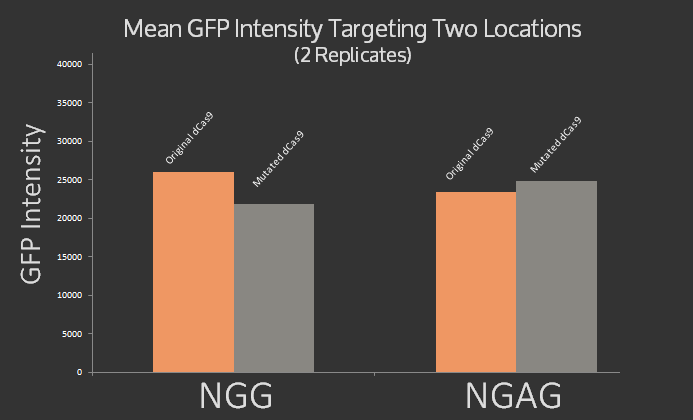
| + | The results are rather inconclusive, as can be seen even more clearly from the bar graph of the data, shown below. It was expected that the original dCas9 would target NGG but not NGAG, while the modified dCas9 would target NGAG but not NGG. Instead roughly the same signal was measured for both dCas9 variants at both target sites. | ||
| + | </p> | ||
| + | <figure> | ||
| + | <img src="/wiki/images/9/95/Waterloo_pamflexlabresults.png" alt="Bar graphs showing results from dCas9 targeting" /> | ||
| + | <figcaption>Mean RFP Intensity after targeting an NGAG and NGG PAM site (using different complementary sgRNA sequences) with both dCas9 and the modified dCas9 variant.</figcaption> | ||
| + | </figure> | ||
| + | </section> | ||
| + | <section id="references" title="References"> | ||
| + | <h2>References</h2> | ||
| + | <ol id="reflist"> | ||
| + | </ol> | ||
| + | </section> | ||
</div> | </div> | ||
</html> | </html> | ||
{{Waterloo_Footer}} | {{Waterloo_Footer}} | ||
Latest revision as of 23:55, 20 November 2015
dCas9 Modification
Introduction
One of our goals this year was to engineer a version of the Cas9 protein that could recognize different protospacer adjacent motifs (PAMs) other than the regular NGG. Kleinstiver et al. recently published data on Cas9 mutants that effectively cut unique PAM sites. A specific EQR variant in the publication recognized a NGAG PAM site after three different amino acids in the original Cas9 were changed. The EQR variant includes the changes D1135E, R1335Q, and T1337R. Our goal was to try and replicate these mutations but instead with dCas9, which is commonly used since it represses a gene rather than cutting one. Target sites were designed in promoters of green fluorescent protein (GFP) with both an NGG PAM sequence and an NGAG PAM sequence.
Experimental Design
All of our experiments to study GFP expression with and without addition of dCas9 were performed on the Amnis Image Stream machine (Amnis 2015). The Image Stream is a fluorescence microscope combined with a flow cytometer, letting one to perform a quantitative method of fluorescence detection while visualizing the actual cells. By measuring an average fluorescence intensity of cells, we can determine the average intensity of a cell population.
Our first step was to create the EQR variant in dCas9 by using site directed mutagenesis. This was done on a dCas9 plasmid ordered from AddGene. Two other plasmids were constructed with both a gene producing GFP and an sgRNA target recognizing either an NGG or an NGAG PAM site on the GFP promoter J23101.
The first experiment that was performed was to co-transform a plasmid containing both GFP and the sgRNA targeting the GFP promoter into the same cell as dCas9. This was a control experiment that will explain that the mutated EQR dCas9 performs worse than the original dCas9 when targeting an NGG PAM site.

The next experiment involved transforming the plasmid with GFP and an sgRNA targeting a region in the GFP promoter with an NGAG PAM site into E. coli. This was also co-transformed with both the orignial and the mutated versions of dCas9. This time, we expect to see the mutated EQR dCas9 perform better than the original in terms of repression of GFP.

Constructs

- GFP with NGAG sgRNA target
- Target plasmid with GFP sgRNA
- Mutated dCas9
- dCas9 from AddGene
Results
The EQR muatated dCas9 was successfully constructed using the QuickChange protocol twice and sending for sequencing. Co-transformations were also accomplished and cells were analyzed using Flow cytometry.
NGG PAM site


NGAG PAM site


These graphs show the results of the mean of the GFP intensity when the co-transformed cells were run on flow cytometry. A lower mean points to the result that the dCas9 is more effective at repressing expression of GFP. These results are inconclusive and we need to repeat the experiment multiple times to understand the effect of the EQR mutations on dCas9.
The results are rather inconclusive, as can be seen even more clearly from the bar graph of the data, shown below. It was expected that the original dCas9 would target NGG but not NGAG, while the modified dCas9 would target NGAG but not NGG. Instead roughly the same signal was measured for both dCas9 variants at both target sites.