Difference between revisions of "Team:SF Bay Area DIYBio/Results"
MariaChavez (Talk | contribs) |
|||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{SF_Bay_Area_DIYBio}} | {{SF_Bay_Area_DIYBio}} | ||
| − | + | __NOTOC__ | |
| + | ==Accomplishments== | ||
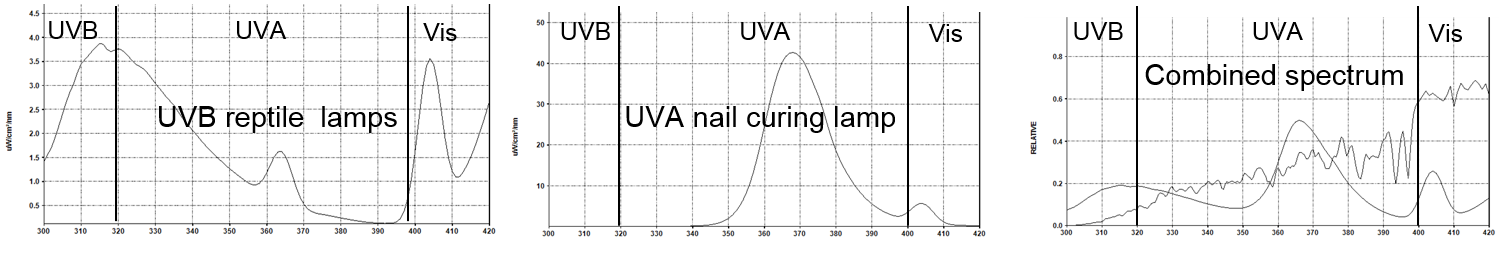
| + | ===Researched UV sources and built UV exposure rig to mimic solar UV=== | ||
| + | *<font size=2>UVA: 320-400nm. “Tanning” wavelengths. Long-term free radical damage | ||
| + | *UVB: 280-320nm. Causes sunburn and direct DNA damage | ||
| + | *UVC: 100-280nm. Rapid skin and retinal damage (e.g.: germicidal UV in BSC)</font> | ||
| − | + | We want to mimic solar UV: broad-band UVA+UVB, but not UVC! After testing many UV sources, we settled on: | |
| + | *<font size=2>UVB basking lamp for pet reptiles | ||
| + | *UVA nail curing lamp</font> | ||
| − | + | [[File:Spectra.png|950px]] | |
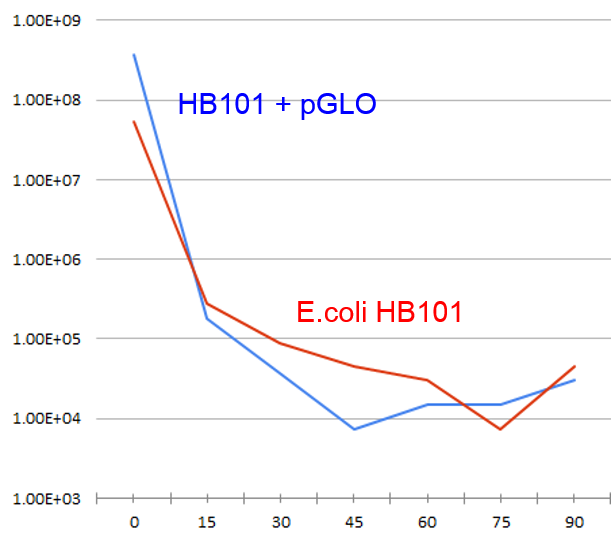
| − | + | ===Determined exposure level needed for >4 log decrease in CFU=== | |
| + | UV kill curve was generated for E.coli HB101, to establish baseline exposure for Directed Evolution | ||
| − | + | We also tested HB101 + pGLO plasmid, to check if GFP has a protective effect. | |
| − | We also tested HB101 + pGLO plasmid, to check if GFP has a protective effect. | + | |
| + | [[File:Kill Curve results.png|500px]] | ||
| + | |||
| + | ===Demonstrated GFP is NOT an effective UV protectant for E.coli=== | ||
4 log decrease in viable cells after 75min for HB101, 30min for pGLO | 4 log decrease in viable cells after 75min for HB101, 30min for pGLO | ||
| + | |||
If anything, pGLO has a negative effect on UV resistance! | If anything, pGLO has a negative effect on UV resistance! | ||
| − | < | + | |
| − | </ | + | ===Synthesized and transformed A. variabilis shinorine biosynthesis genes=== |
| + | |||
| + | ===Submitted one gene (Ava_3856) to Parts Registry=== | ||
| + | |||
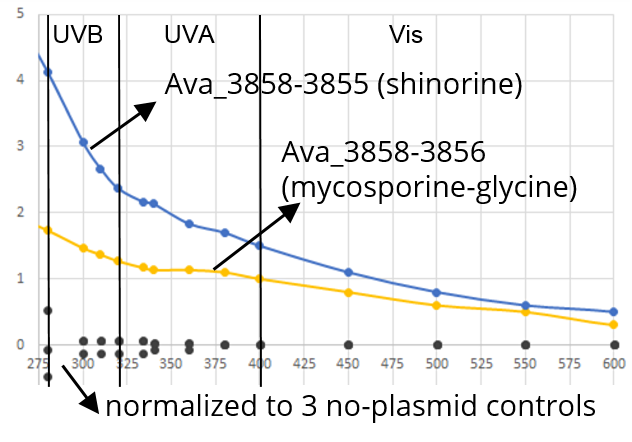
| + | ===Demonstrated that we produced UV absorbing compounds=== | ||
| + | MAA’s can be extracted with methanol: | ||
| + | *<font size=2>Grow 20ml culture overnight in TSB | ||
| + | *Spin down & wash in saline 2x | ||
| + | *Extract in 2ml methanol at 4C overnight | ||
| + | *Pellet at 10,000rpm, collect supernatant | ||
| + | *Collect UV absorption spectrum: | ||
| + | **E. coli HB101 (control) | ||
| + | **HB101+Ava_3858-3855 (full shinorine pathway) | ||
| + | **HB101+Ava_3858-3856 (up to mycosporine-glycine only)</font> | ||
| + | |||
| + | [[File:UV Absorption Curve.png|500px]] | ||
| + | |||
| + | Yay - we’re producing UV absorbing compounds! Further analysis will have to wait until we get our HPLC up and running... | ||
Latest revision as of 13:14, 21 November 2015
Accomplishments
Researched UV sources and built UV exposure rig to mimic solar UV
- UVA: 320-400nm. “Tanning” wavelengths. Long-term free radical damage
- UVB: 280-320nm. Causes sunburn and direct DNA damage
- UVC: 100-280nm. Rapid skin and retinal damage (e.g.: germicidal UV in BSC)
We want to mimic solar UV: broad-band UVA+UVB, but not UVC! After testing many UV sources, we settled on:
- UVB basking lamp for pet reptiles
- UVA nail curing lamp
Determined exposure level needed for >4 log decrease in CFU
UV kill curve was generated for E.coli HB101, to establish baseline exposure for Directed Evolution
We also tested HB101 + pGLO plasmid, to check if GFP has a protective effect.
Demonstrated GFP is NOT an effective UV protectant for E.coli
4 log decrease in viable cells after 75min for HB101, 30min for pGLO
If anything, pGLO has a negative effect on UV resistance!
Synthesized and transformed A. variabilis shinorine biosynthesis genes
Submitted one gene (Ava_3856) to Parts Registry
Demonstrated that we produced UV absorbing compounds
MAA’s can be extracted with methanol:
- Grow 20ml culture overnight in TSB
- Spin down & wash in saline 2x
- Extract in 2ml methanol at 4C overnight
- Pellet at 10,000rpm, collect supernatant
- Collect UV absorption spectrum:
- E. coli HB101 (control)
- HB101+Ava_3858-3855 (full shinorine pathway)
- HB101+Ava_3858-3856 (up to mycosporine-glycine only)
Yay - we’re producing UV absorbing compounds! Further analysis will have to wait until we get our HPLC up and running...