Difference between revisions of "Team:TJU/Results"
| Line 161: | Line 161: | ||
<p>2. Glucose consumption curve</p> | <p>2. Glucose consumption curve</p> | ||
<div class="kuang" style="width:770px"> | <div class="kuang" style="width:770px"> | ||
| − | </br><a href="https://static.igem.org/mediawiki/2015/ | + | </br><a href="https://static.igem.org/mediawiki/2015/1/16/Result_2%281%29.png |
| − | " target="_blank" ><img src= "https://static.igem.org/mediawiki/2015/ | + | " target="_blank" ><img src= "https://static.igem.org/mediawiki/2015/1/16/Result_2%281%29.png |
" width="750" alt=""/></a> | " width="750" alt=""/></a> | ||
<div id="Enlarge"> | <div id="Enlarge"> | ||
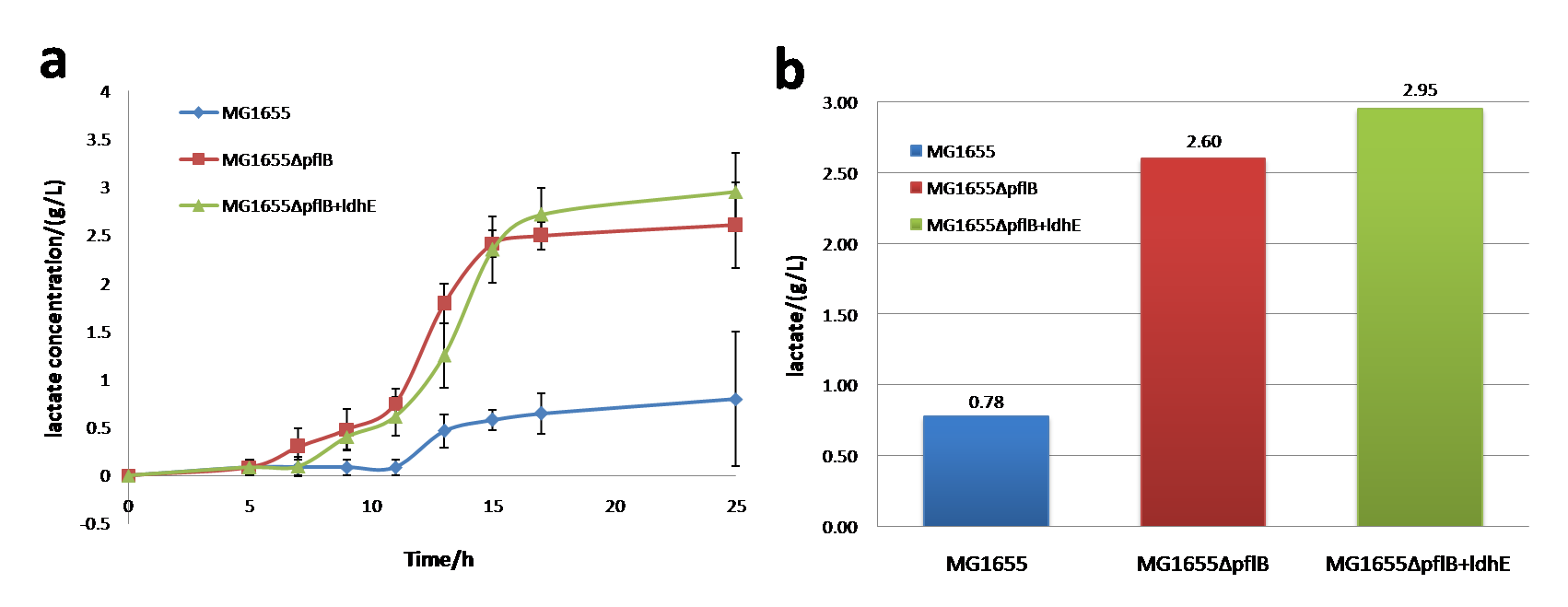
<p> <b>Figure 2.</b> <span style="font-size: 14px"> a: The lactate production curve in anaerobic condition (M9 medium)b: The lactate production curve in anaerobic condition (M9 medium)</span> | <p> <b>Figure 2.</b> <span style="font-size: 14px"> a: The lactate production curve in anaerobic condition (M9 medium)b: The lactate production curve in anaerobic condition (M9 medium)</span> | ||
| − | <a href="https://static.igem.org/mediawiki/2015/ | + | <a href="https://static.igem.org/mediawiki/2015/1/16/Result_2%281%29.png |
" target="_blank"><img src=" https://static.igem.org/mediawiki/2013/9/90/Enlarge.jpg " width="20" height="20" align="right" alt="" /></a></p></div></div></br> | " target="_blank"><img src=" https://static.igem.org/mediawiki/2013/9/90/Enlarge.jpg " width="20" height="20" align="right" alt="" /></a></p></div></div></br> | ||
<p>The anaerobically consumption rate keeps the nearly same among MG1655, MG1655Δ<span style="font-style: italic">pflB</span> and MG1655Δ<span style="font-style: italic">pflB</span>+<span style="font-style: italic">ldhE</span>.</p></br> | <p>The anaerobically consumption rate keeps the nearly same among MG1655, MG1655Δ<span style="font-style: italic">pflB</span> and MG1655Δ<span style="font-style: italic">pflB</span>+<span style="font-style: italic">ldhE</span>.</p></br> | ||
Revision as of 17:03, 17 September 2015

Results
1 Lactate producing system
1.
Under anaerobic conditions, wild-type MG1655 has the same growth rate as MG1655 with ΔpflB and MG1655 with ΔpflB together with the insertion of ldhE keeps relatively lower growth rate than the two mentioned before.
2. Glucose consumption curve
The anaerobically consumption rate keeps the nearly same among MG1655, MG1655ΔpflB and MG1655ΔpflB+ldhE.
3和4乳酸生成曲线 lactate production curveIn anaerobic environment, lactate production of MG1655, MG1655 ΔpflB and MG1655 ΔpflB+ldhE has large differences between each other. Notably, knocking out of pflB will increase lactate generation up to ~2 g/L and the amount produced by MG1655ΔpflB+ldhE is slightly greater than MG1655ΔpflB.
Conclusion: knocking out of pflB can dramatically improve the lactate production. Besides, ldhE also plays an important role in lactate increase. We improve our utilization of carbon sources by constructing a high-yield strain with M9 medium under the same concentrations of glucose.
2 Flavins producing system
As shown in figure 3, E. coli BL21 with EC10 reaches a final yield of 17 mg/L in tube cultivation. E. coli BL21Δpgi+EC10 reaches 33 mg/L. E. coli BL21+EC10* reaches 90 mg/L.
3 Co-culture MFC -- Labor Division
Figure 9: Among optimized shewanella, shewanella+MG1655 and Shewanella+ΔplfB ldhE+Rf02S, the three-strain coculture system reached a relatively higher potential that kept around 350 mV in 80 hours.
Conclusion: Shewanella+ΔplfB ldhE+Rf02S has been going through a preferable optimization. It led to the potential of 300 mV in 80 hours under the condition of 2 g/L glucose without any supplementary. So, it realized a higher and more endurable power generation when taking glucose as carbon sources in Shewanella.
Figure 10: The potential and time of duration in Shewanella+ΔplfB ldhE+B.Subtilis system can be raised into 511 mV and 80 h, respectively, resulted in a greater generation amount.
Figure 11:
The potential produce by Shewanella+ΔplfB ldhE+B.Subtilis is greater than Shewanella+ΔplfB ldhE+Rf02S for 176 mV. Cocultured Shewanella, E.coli and B.subtilis can achieve a better job division that can reduce the competition between two kinds of zymophyte since B,subtilis can anaerobically metabolize by using KNO3. It significantly improve the commensalism relations between three species.
Figure 12
(1) A series of voltage brought by different external resistance from Shewanella+ΔplfB ldhE+Rf02S (2) The relations between current density and voltage is well suited to polarization curve while the power curve can represent the relations between output power density and current density. Besides, the highest power density can reach to 10 mW/m2.Figure 13
(1) A series of voltage brought by different external resistance from Shewanella+ΔplfB ldhE+B.Subtilis
(2) The relations between current density and voltage is well suited to polarization curve while the power curve can represent the relations between output power density and current density. Besides, the highest power density can reach to 17 mW/m2.
Figure 14
The comparison of polarization curve and power curve among shewanella, shewanella+MG1655, shewanella+ΔplfB ldhE+Rf02S. It is obvious that the power output in shewanella+ΔplfB ldhE+Rf02S is far more bigger than the controled group.Figure 15
The comparison of polarization curve and power curve among shewanella, shewanella+MG1655 and Shewanella+ΔplfB ldhE+B.Subtilis. We can apparently observed that the power output of Shewanella+ΔplfB ldhE+B.Subtilis is greater than the controled group.
Figure 16
The comparison of polarization curve and power curve among Shewanella+ΔplfB ldhE+B.Subtilis and Shewanella+ΔplfB ldhE+Rf02S. It has been shown in power curve that Shewanella+ΔplfB ldhE+B.Subtilis has a higher output than Shewanella+ΔplfB ldhE+Rf02S, which indicates that three-strain system can generate higher electricity and have a better MFC performance, in return, have a promising application.
Future work
Proteolysis system
During the experiment, our MFC in anaerobic condition has been found to reach an unexpected lower pH that showed harmful to bacterial consortium. On account of improving the rate of survival, we designed a “Sensing- Regulating” system to consume the accumulated lactate and keep the environment at a proper acid level. Our system belongs to proteomic regulation, which contains pH sensing system and orthogonal proteolysis system for self-regulating. The delayed effect within our sensing system brings us a more precise depict of possible result that the pH may oscillate at a certain range.
1 Sensing
Every genus of cell has an optimum range of pH to live and Shewanella along with E.coli is no exception. Previous research has identified that Shewanella can maintain a maximal population density when pH keeps above 6.2 while E.coli can survive in 5.6~8.1.[1] As an attempt to develop the superior sensing mechanism, we screen out promoter 170 (P170) and regulatory protein RcfB and introduce them into E.coli for adaption.
P170, a strongly acid-inducible promoter from Lactococcus lactis, is up-regulated at low pH during the transition to stationary phase.[2] The trans-acting protein RcfB, however, is involved in basal activity of P170 and is also essential for pH induction. The protein RcfB, upon activation by lactate, bind a DNA recognition motif within a promoter region and activates transcription.[3] When the cells are exposed to acid environments, the RcfB activation by an ‘acid’ signal allows its binding to the ACiD-box, resulting in transcription activation.[3]
To test the idea that P170 is sensitive to acidity under the regulation of RcfB, we construct RFP gene into circuit containing P170 and RcfB. The RFP is expressed on the basis of P170 activation and RcfB which functions as a novel regulatory protein however, is controlled by a constitutive promoter J23100. High level of P170 activity required both RcfB and acidic conditions. Therefore, with the constitutive expression of RcfB, once the pH reaches the threshold that usually ranges from pH 6.0 to pH 6.5, P170 will be induced so that RFP can reflect red fluorescence under UV detection. Although RFP verification test is well developed, other functional genes are also replace RFP gene as a strategy for application extension. In our later experiment, we substitute RFP for T7 polymerase for regulatory purpose.
2 Regulating
After activated sensing system which determines the pH threshold to suppress bacterial population density, we need to figure out how to retrieve to the proper acidity for living. Here comes our regulating system for acid recovery.
The ssrA tag sequence (mf-ssrA tag) and the Lon protease (mf-Lon) from Mesoplasma florum are made up of the crucial elements of a protein degradation system. Experiments has been identified that mf-ssrA tag is efficiently recognized by mf-Lon where the tag appends to C terminus of native or denatured proteins resulted in their rapid proteolysis by mf-Lon.[4] Besides, degradation system based on the Gram-positive M. florum tmRNA system does not rely on host degradation systems and can function in a wide range of bacteria, which makes it an adaptive method in variety. Here, we use this proteolysis system for lactate regulation and keep it in a oscillatory value of acidity.
Specifically, we utilize the effect of mf-Lon as well as ldhE bearing with tag under the control T7 promoter and J23100 promoter respectively, to degrade the overmuch lactate. We also aware that T7 promoter activation requires the combination of T7 RNA polymerase which is extremely promoter-specific and transcribes only DNA downstream of a T7 promoter.[5] According to the extensive study of our ‘sensing’ system, we substitute the RFP for T7 polymerase hoping that lower pH stage can activate T7 polymerase activation which then combines with T7 promoter for acting and mf-Lon, in this way, also expresses. It turns out that ldhE togethered with mf-ssrA tag can be degraded by cytoplasmic mf-Lon. So the pH can maintain in a proper level for cells’ living.
(实验表征结果和说明)Notably, the proteolysis system has potential to apply in many fields. As a strategy to make it a toolbox for further research, we use different tags to test the strength of its proteolysis. With the reflection of red fluorescence, we are able to detect mf-Lon activity on various tags based on the distinguish value of UV detection. This toolbox is intended to be a fast and convenient method for prospective proteolysis option and degradation research.
(实验表征结果和说明)References
[1] Min C, Chung H, Choi W, et al. Linear correlation between inactivation of E. coli and OH radical concentration in TiO2 photocatalytic disinfection.[J]. Water Research, 2004, 38(4):1069–1077. [2] Madsen S M, Arnau J, Vrang A, et al. Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis.[J]. Molecular Microbiology, 1999, 32(1):75-87. [3] Madsen, Søren M, Hindré, Thomas, Le Pennec, Jean‐Paul, et al. Two acid‐inducible promoters from Lactococcus lactis require the cis‐acting ACiD‐box and the transcription regulator RcfB[J]. Molecular Microbiology, 2005, 56(3):735-746. [4] Eyal G, Sauer R T. Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease.[J]. Proceedings of the National Academy of Sciences, 2008, 105(42):16113-16118. [5] Martin C T, Esposito E A, Theis K, et al. Structure and function in promoter escape by T7 RNA polymerase.[J]. Prog Nucleic Acid Res Mol Biol, 2005, 80(80):323–347.
E-mail: ggjyliuyue@gmail.com |Address: Building No.20, No.92 Weijin road, Tianjin University, China | Zip-cod: 300072
Copyright 2015@TJU iGEM Team