|
|
| Line 1: |
Line 1: |
| | <html> | | <html> |
| − | <head> | + | <head> |
| − | <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> | + | <meta charset="utf-8"> |
| − | <title></title> | + | <meta http-equiv="X-UA-Compatible" content="IE=edge"> |
| − | <link href="https://2015.igem.org/Team:NEFU_China/navcss?action=raw&ctype=text/css" rel="stylesheet" type="text/css" />
| + | <meta name="viewport" content="width=device-width, initial-scale=1"> |
| − | <link href="https://2015.igem.org/Team:NEFU_China/btop?action=raw&ctype=text/css" rel="stylesheet" type="text/css" /> | + | <!-- The above 3 meta tags *must* come first in the head; any other head content must come *after* these tags --> |
| − | <script type="text/javascript" src="//code.jquery.com/jquery-1.11.3.min.js"></script>
| + | <title>Flight iGEM</title> |
| − | <script type="text/javascript" src="https://2015.igem.org/Team:NEFU_China/navjs?action=raw&ctype=text/javascript"></script>
| + | |
| − | <style>
| + | |
| − | #content{
| + | |
| − | background:none;
| + | |
| − | text-align:justify;
| + | |
| − | }
| + | |
| − | .mw-content-ltr ul, .mw-content-rtl .mw-content-ltr ul{
| + | |
| − | margin: 0;
| + | |
| − | }
| + | |
| − | #content img{magin:auto;}
| + | |
| − | </style>
| + | |
| − | <link href="https://2015.igem.org/Team:NEFU_China/cleariGEM?action=raw&ctype=text/css" rel="stylesheet" /> </head>
| + | |
| − | <body style="background:url(https://static.igem.org/mediawiki/2015/e/e3/NEFU_China_3F0097CC-EE0E-4A38-8313-BD08C32A5571.gif) no-repeat top left #00181f;background-attachment: fixed;"> | + | |
| − | <!--nav star-->
| + | |
| − | <script type="text/javascript" src="https://2015.igem.org/Team:NEFU_China/nav?action=raw&ctype=text/javascript"></script> | + | |
| − | <script type="text/javascript" src="https://2015.igem.org/Team:NEFU_China/btopjs?action=raw&ctype=text/javascript"></script>
| + | |
| − | <!--nav end-->
| + | |
| | | | |
| − | <div class="s2_1box" style="background:none;color:white;"> | + | <!-- Bootstrap --> |
| − | <p><strong><span style="font-size:16px"><span style="font-family:arial,helvetica,sans-serif">7. pHY300PLK-plsr-blue-T</span></span></strong></p> | + | <link href="https://maxcdn.bootstrapcdn.com/bootstrap/3.3.5/css/bootstrap.min.css" rel="stylesheet"> |
| | + | <link href="https://2015.igem.org/Team:NEFU_China/fullPagecss?action=raw&ctype=text/css" rel="stylesheet"> |
| | | | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1. Molecular biology techniques: SOE (Splicing by overlap extension) PCR</span></p> | + | <!-- HTML5 shim and Respond.js for IE8 support of HTML5 elements and media queries --> |
| | + | <!-- WARNING: Respond.js doesn't work if you view the page via file:// --> |
| | + | <!--[if lt IE 9]> |
| | + | <script src="https://oss.maxcdn.com/html5shiv/3.7.2/html5shiv.min.js"></script> |
| | + | <script src="https://oss.maxcdn.com/respond/1.4.2/respond.min.js"></script> |
| | + | <![endif]--> |
| | + | <!--<link href='https://http://fonts.googleapis.com/css?family=Open+Sans' rel='stylesheet' type='text/css'>--> |
| | + | <style type="text/css"> |
| | + | body{ |
| | + | text-align: justify; |
| | + | font-size: 16px; |
| | + | font-family: 'Open Sans', sans-serif; |
| | + | } |
| | + | h1, h2, h3, h4, h5, h6{ |
| | + | border-bottom: none; |
| | + | } |
| | + | #bodyContent h1, #bodyContent h2{ |
| | + | margin-bottom: 0; |
| | + | } |
| | + | .section h2{ |
| | + | text-align: center; |
| | + | padding-bottom: 5px; |
| | + | position: relative; |
| | + | line-height: 80px; |
| | + | color: white; |
| | + | } |
| | + | .section h2 em{ |
| | + | display: block; |
| | + | border-width: 10px; |
| | + | position: absolute; |
| | + | bottom: -10px; |
| | + | left: 50%; |
| | + | border-style: solid dashed dashed; |
| | + | border-color: transparent transparent #F7931E transparent; |
| | + | font-size: 0; |
| | + | line-height: 0; |
| | + | } |
| | + | .section .navbar { |
| | + | margin-bottom: 120px; |
| | + | position: absolute; |
| | + | top: 0; |
| | + | width: 100%; |
| | + | } |
| | + | .section .navbar h2{ |
| | + | margin-top:0; |
| | + | } |
| | + | .logo-container{ |
| | + | height: 160px; |
| | + | background-color: #F7931E; |
| | + | width: 100px; |
| | + | position: absolute; |
| | + | left: 20px; |
| | + | } |
| | + | .logo { |
| | + | display: block; |
| | + | font-size: 15px; |
| | + | margin-top: 125px; |
| | + | text-align: center; |
| | + | color: white; |
| | + | } |
| | + | @media (max-width: 768px) { |
| | + | .logo-container { |
| | + | display: none; |
| | + | } |
| | + | } |
| | + | .sub-container{ |
| | + | padding-top: 45px; |
| | + | } |
| | + | .sub-container .msg { |
| | + | background-color: #f2f2f2; |
| | + | position: relative; |
| | + | min-height: 250px; |
| | + | padding: 15px; |
| | + | display: block; |
| | + | } |
| | + | .chart-msg { |
| | + | padding: 15px; |
| | + | padding-left: 30px; |
| | + | } |
| | + | .sub-container .img{ |
| | + | display: block; |
| | + | } |
| | + | .sub-container .msg em { |
| | + | position: absolute; |
| | + | border-width: 10px; |
| | + | font-size: 0; |
| | + | line-height: 0; |
| | + | } |
| | + | .msg .right{ |
| | + | top: 50%; |
| | + | right: -20px; |
| | + | border-style: dashed solid solid dashed; |
| | + | border-color: transparent transparent transparent #f2f2f2; |
| | + | } |
| | + | .msg .left{ |
| | + | top: 50%; |
| | + | left: -20px; |
| | + | border-style: dashed solid solid dashed; |
| | + | border-color: transparent #f2f2f2 transparent transparent; |
| | + | } |
| | + | .msg .up{ |
| | + | top: -20px; |
| | + | left: 50%; |
| | + | border-style: dashed solid solid dashed; |
| | + | border-color: transparent transparent #f2f2f2 transparent; |
| | + | } |
| | + | .fp-controlArrow .fp-prev{ |
| | + | border-color: transparent gray transparent transparent; |
| | + | } |
| | + | .fp-controlArrow .fp-next{ |
| | + | border-color: transparent transparent transparent gray; |
| | + | } |
| | + | #section-1{ |
| | + | background: -webkit-gradient(linear, top left, bottom left, from(#4bbfc3), to(#7baabe)); |
| | + | background: -webkit-linear-gradient(#4BBFC3, #7BAABE); |
| | + | background: linear-gradient(#4BBFC3,#7BAABE); |
| | + | } |
| | + | .show-sec{ |
| | + | background: -webkit-gradient(linear, top left, bottom left, from(#7baabe), to(#969ac6)); |
| | + | background: -webkit-linear-gradient(#7BAABE, #969AC6); |
| | + | background: linear-gradient(#7BAABE,#969AC6); |
| | + | } |
| | + | .section .himg{ |
| | + | width:100%; |
| | + | } |
| | + | </style> |
| | + | <link href="https://2015.igem.org/Team:NEFU_China/cleariGEM?action=raw&ctype=text/css" rel="stylesheet" /> </head> |
| | + | <body> |
| | + | <div id="fullpage"> |
| | + | <div class="section" id="section-1"> |
| | + | <nav class="navbar navbar-inverse"> |
| | + | <div class="logo-container"><span class="logo">Flight iGEM</span></div> |
| | + | <h2>Facts<em></em></h2> |
| | + | </nav> |
| | + | <div class="container"> |
| | + | <div class="row"> |
| | + | <div class="sub-container"> |
| | + | <h2 style="text-align:center;">Attitudes towards creating a wiki</h2> |
| | + | <div class="col-xs-6 col-md-6"> |
| | + | <canvas id="slide-canvas-1"></canvas> |
| | + | </div> |
| | + | <div class="col-xs-6 col-md-6 chart-msg"> |
| | + | <p>According to a survey conducted by us, we found that most teams agree the thesis that creating wiki is a difficult or tedious work to finish with high quality, especially when time is very limited. So, we’d like to make some changes.</p> |
| | | | |
| − | <p><span style="font-size:16px"><strong><span style="font-family:arial,helvetica,sans-serif">A. Primary PCR reaction</span></strong></span></p> | + | <p>*You can see all raw data by click <a href="http://www.wenjuan.com/r/QF3QnyK?pid=55f01be9f7405b0c34569df1&vcode=d3452b2f6d3f5f4e009805e3520d4512">this</a>.</p> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="section show-sec" id="section"> |
| | + | <nav class="navbar navbar-inverse"> |
| | + | <div class="logo-container"><span class="logo">Flight iGEM</span></div> |
| | + | <h2>Flight iGEM Magic<em></em></h2> |
| | + | </nav> |
| | + | <div class="slide" id="slide1"> |
| | + | <div class="container"> |
| | + | <div class="row"> |
| | + | <div class="sub-container"> |
| | + | <div class="col-xs-6 col-md-6" style="text-align:center;"><img src="https://static.igem.org/mediawiki/2015/6/64/NEFU_China_0E792055-5CA4-4CE3-8638-93AC5988AC34.png" /></div> |
| | | | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Segment1-plsr</span></strong></p> | + | <div class="col-xs-6 col-md-6 msg"> |
| | + | <p>About half of teams we surveyed hold the opinion that creating a wiki requires superior skills in programming (29/56), which lead them feel hard about it. We sincerely invite you to click the right arrow and discover what giant changes Flight iGEM can make.</p> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="slide"> |
| | + | <div class="container"> |
| | + | <div class="raw"> |
| | + | <div class="sub-container"> |
| | + | <div class="col-xs-6 col-md-6"><img class="himg" alt="" src="https://static.igem.org/mediawiki/2015/6/61/NEFU_China_FABE4DBA-CD83-486D-AAC2-E4B3949D76D8.png" /></div> |
| | | | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/f3/NEFU_China_31952388-2D19-4FA8-8DBD-63EDA1E2CB4F.png" style="height:344px; width:700px" /></p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h1>Step 1</h1> |
| | | | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Segment2- Blue-T</span></strong></p> | + | <p>To create a wiki, a HTML template is necessary neither with Flight iGEM nor without Flight iGEM.</p> |
| | + | </div> </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="slide"> |
| | + | <div class="container"> |
| | + | <div class="raw"> |
| | + | <div class="sub-container"> |
| | + | <h1>Step 2</h1> |
| | | | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/f7/NEFU_China_B4879185-4C26-4FC5-A133-5F5056B51D02.png" style="height:399px; width:700px" /></p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>Without Flight iGEM</h2> |
| | + | <img alt="" class="himg" src="https://static.igem.org/mediawiki/2015/3/30/NEFU_China_60AF68F6-696F-4A98-844E-2023C8011191.png" /> |
| | + | <p>Once the template is done, other teammates can start to summarize the project, record experiments’ data by using their Word or iWork.</p> |
| | + | </div> |
| | | | |
| − | <p> </p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>With Flight iGEM</h2> |
| | + | <img alt="" class="himg" src="https://static.igem.org/mediawiki/2015/6/6f/NEFU_China_75095857-0A36-4862-8293-8993E03284D5.png" />Flight iGEM requires you and your teammates to work online. We use CKEditor(a free, Open Source HTML text editor) to set up edit environment like Word or iWork.</div> </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="slide"> |
| | + | <div class="container"> |
| | + | <div class="raw"> |
| | + | <div class="sub-container"> |
| | + | <h1>Step 3</h1> |
| | | | |
| − | <p style="margin-left:18pt; text-align:center"><span style="color:#FF0000"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/f/f9/NEFU_China_FAFBCF0A-D050-461A-A4A2-825CD2A6B4E2.png" style="height:150px; width:150px" /></span></span></p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>Without Flight iGEM</h2> |
| | + | <img class="himg" src="https://static.igem.org/mediawiki/2015/3/3c/NEFU_China_53ABE671-6E9D-4A6A-A6CA-6C58897625F2.png" /> |
| | + | <p>Without Flight iGEM, you need to upload all images and other media files which involved in your articles to the iGEM server manually.</p> |
| | + | </div> |
| | | | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 34. 1. Marker (DL2000) ; 2. Blue-T (878bp) ; 3. plsr (250bp)</span></p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>With Flight iGEM</h2> |
| | + | <img class="himg" src="https://static.igem.org/mediawiki/2015/0/09/NEFU_China_7C064076-7096-4A36-918C-6602AC0862D3.png" /> |
| | + | <p>If you use Flight iGEM, all media files(especially the images) which you insert into the article will be automatically upload to iGEM server.</p> |
| | + | </div> </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="slide"> |
| | + | <div class="container"> |
| | + | <div class="raw"> |
| | + | <div class="sub-container"> |
| | + | <h1>Step 4</h1> |
| | | | |
| − | <p style="text-align:center"> </p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>Without Flight iGEM</h2> |
| | + | <img class="himg" src="https://static.igem.org/mediawiki/2015/e/e6/NEFU_China_DEE6107D-1149-460D-A6CA-F9A0FCEEBE66.png" /> |
| | + | <p>You need to assign the content which converted from Word or iWork documents to the template. In this step, there will be numerous words and sentences that will be copied and pasted, which cost a lot of time and memory.</p> |
| | + | </div> |
| | | | |
| − | <p style="text-align:center"> </p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>With Flight iGEM</h2> |
| | + | <img class="himg" src="https://static.igem.org/mediawiki/2015/d/d5/NEFU_China_8EA8F25C-716E-480F-9DFC-B3B0B58DD2B0.png" /> |
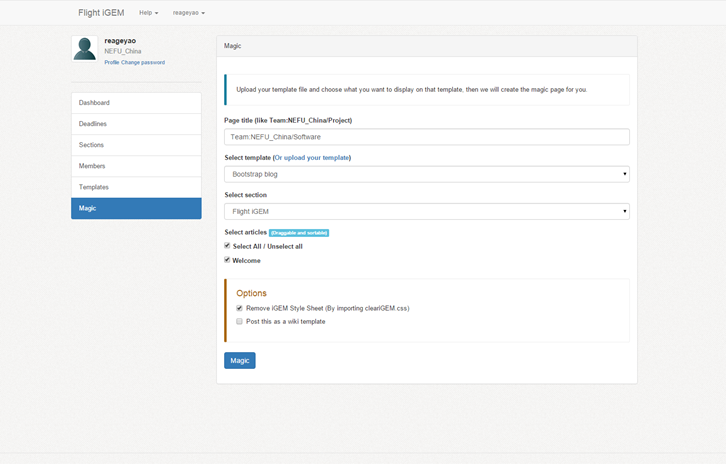
| | + | <p>Flight iGEM can assign contents with template automatically, all you need to do is to select what you want to display, and then click 'Magic'.</p> |
| | + | </div> </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="slide"> |
| | + | <div class="container"> |
| | + | <div class="raw"> |
| | + | <div class="sub-container"> |
| | + | <h1>Step 5</h1> |
| | | | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">B. Overlapping and elongation</span></strong></p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>Without Flight iGEM</h2> |
| | + | <img class="himg" src="https://static.igem.org/mediawiki/2015/0/05/NEFU_China_419F93B3-DBB5-46EF-A785-05A0D702C8F0.png" /> |
| | + | <p>After you merged this two page, there are you should upload the "code soup" to the iGEM server, and this is the final step of the whole procedure.</p> |
| | + | </div> |
| | | | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/0/0b/NEFU_China_6F230DAC-C5D6-4BDB-88AA-02DA124DC03F.png" style="height:278px; width:700px" /></p> | + | <div class="col-xs-6 col-md-6"> |
| | + | <h2>With Flight iGEM</h2> |
| | + | <img class="himg" src="https://static.igem.org/mediawiki/2015/6/6a/NEFU_China_E5C6420A-4FEF-4898-BDED-AFCAEB7E36AF.png" /> |
| | + | <p>Flight iGEM will upload your pages to the iGEM server after step 4, and the only thing you need to do is to visit your target page and check if the page is exactly what you want.</p> |
| | + | </div> </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="section show-sec"> |
| | + | <nav class="navbar navbar-inverse"> |
| | + | <div class="logo-container"><span class="logo">Flight iGEM</span></div> |
| | + | <h2>Online Collaboration<em></em></h2> |
| | + | </nav> |
| | + | <div class="container"> |
| | + | <div class="row"> |
| | + | <div class="sub-container"> |
| | + | <div class="col-xs-6 col-md-6" style="text-align:center;"><img src="https://static.igem.org/mediawiki/2015/3/38/NEFU_China_93E048E2-4DF1-4B5C-B5A7-AAECD232B31F.png" /></div> |
| | + | <div class="col-xs-6 col-md-6 msg"> |
| | + | <p>A typical iGEM wiki can be divided into no less than 8 sections (like the home page, project, safety, team, and so on). Under each section, there are a lot of issues which you want to present. Just like the ‘Project’ section, you may need to tell others about your background information, design, etc. Tedious as it is, we, the same as other 52 teams (52/56), believe that online collaboration can improve the efficiency of creating a wiki. Flight iGEM provides you an online platform, in which you can collaborate with your teammates, so every teammate can create page and edit them at any time or anywhere. To avoid version conflict(covered by others’ edition), we mimic the concept of SVN. You can click <a href="#" data-toggle="modal" data-backdrop="" data-target="#vcModal">me</a> to watch a short video which explains how it works.</p> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="section show-sec"> |
| | + | <nav class="navbar navbar-inverse"> |
| | + | <div class="logo-container"><span class="logo">Flight iGEM</span></div> |
| | + | <h2>Deadline alarm<em></em></h2> |
| | + | </nav> |
| | + | <div class="container"> |
| | + | <div class="row"> |
| | + | <div class="sub-container"> |
| | + | <div class="col-xs-12 col-md-12" style="text-align:center;"><img src="https://static.igem.org/mediawiki/2015/9/9b/NEFU_China_3702A597-6FFB-4715-990E-36F15066FBF3.png" /></div> |
| | | | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">C. Second PCR reaction</span></strong></p> | + | <div class="col-xs-12 col-md-12 msg"> |
| | + | <em class="up"></em> |
| | + | <p>According to our investigation among 65 teams, 18 of them thought there were too many deadlines in iGEM and they might be forgotten easily. Moreover,43 teams surveyed indicated that it would be better if there are certain reminders for the deadlines. As we all know, iGEM is a competition composed of a lot of events. To help users manage their time better, Flight iGEM also covers a deadline reminder. Most noteworthy is that this reminder is able to convert time zones automatically according to the users' geographic positions.</p> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <div class="section show-sec"> |
| | + | <nav class="navbar navbar-inverse"> |
| | + | <div class="logo-container"><span class="logo">Flight iGEM</span></div> |
| | + | <h2>What's for the iGEM Server?<em></em></h2> |
| | + | </nav> |
| | + | <div class="container"> |
| | + | <div class="raw"> |
| | + | <div class="sub-container"> |
| | + | <div class="col-xs-6 col-md-6" style="text-align:center;"><img class="himg" src="https://static.igem.org/mediawiki/2015/d/d8/Queue.png" /></div> |
| | | | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/8/8d/NEFU_China_1E5EE16F-C0AB-4C36-85FB-F6082564BA8C.png" style="height:374px; width:700px" /></p> | + | <div class="col-xs-6 col-md-6 msg"> |
| | + | <p>Considering that 83% teams (54/65) have encountered the situation that the iGEM server got slower or went down when it is close to the freeze of wiki, we decide to linearize all our request. It means if you want to build up a page and post it to iGEM server, you have to queue for a while until people who requested before you finished their query. To speed up all the requests, we create more than ten queues, which we believe is enough to handle them. And we hold the opinion that our platform will contribute a lot to ease the burden of iGEM servers, if it is accepted by other teams.</p> |
| | + | </div> </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <!-- VC Modal --> |
| | + | <div class="modal fade" id="vcModal" tabindex="-1" role="dialog" aria-labelledby="vcModalLabel"> |
| | + | <div class="modal-dialog" role="document"> |
| | + | <div class="modal-content"> |
| | + | <div class="modal-header"> |
| | + | <button type="button" class="close" data-dismiss="modal" aria-label="Close"><span aria-hidden="true">×</span></button> |
| | + | <h4 class="modal-title" id="vcModalLabel">How version control works</h4> |
| | + | </div> |
| | + | <div class="modal-body" style="text-align:center;"> |
| | + | <video width="580" height="360" controls="controls"> |
| | + | <source src="https://static.igem.org/mediawiki/2015/4/49/NEFU_China_vc.ogg" type="video/ogg"> |
| | + | <source src="https://static.igem.org/mediawiki/2015/b/b0/NEFU_China_vc.mp4" type="video/mp4"> |
| | + | Your browser does not support the video tag. |
| | + | </video> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | <!-- jQuery (necessary for Bootstrap's JavaScript plugins) --> |
| | + | <script src="//code.jquery.com/jquery-1.11.3.min.js"></script> |
| | + | <!-- Include all compiled plugins (below), or include individual files as needed --> |
| | + | <script src="https://maxcdn.bootstrapcdn.com/bootstrap/3.3.5/js/bootstrap.min.js"></script> |
| | + | <!--<script src="js/jquery.easings.min.js"></script>--> |
| | + | <script src="https://2015.igem.org/Team:NEFU_China/fullPage?action=raw&ctype=text/javascript"></script> |
| | + | <script src="https://2015.igem.org/Team:NEFU_China/Chartjs?action=raw&ctype=text/javascript"></script> |
| | + | <script type="text/javascript"> |
| | + | $(document).ready(function() { |
| | + | $('#fullpage').fullpage({ |
| | + | anchors: ['intro', 'builder', 'collaboration', 'deadline', '4igem'], |
| | + | navigation: true, |
| | + | navigationPosition: 'right', |
| | + | menu: '#menu', |
| | + | css3: true, |
| | + | scrollingSpeed: 666, |
| | + | navigationTooltips: ['Some facts about creating a wiki', 'Flight iGEM Magic', 'Online Collaboration', 'Deadline', 'For iGEM'] |
| | + | }); |
| | | | |
| − | <p><span style="color:#FF0000"><span style="font-family:arial,helvetica,sans-serif"> <img alt="" src="https://static.igem.org/mediawiki/2015/9/9b/NEFU_China_1759504E-CD10-448E-A49A-81693C20EF31.png" style="height:150px; width:150px" /></span></span></p>
| + | $('#showExamples').click(function(e){ |
| | + | e.stopPropagation(); |
| | + | e.preventDefault(); |
| | + | $('#examplesList').toggle(); |
| | + | }); |
| | | | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 35. 1. Marker (DL2000) ; 2. pLsr-Blue-T (1128bp) ; 3. pLsr (250bp) ; 4. Blue-T (878bp)</span></p>
| + | $('html').click(function(){ |
| | + | $('#examplesList').hide(); |
| | + | }); |
| | | | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | var chart1 = { |
| | + | labels : ['hard', 'tedious', 'not difficult', 'pleasant'], |
| | + | datasets : [ |
| | + | { |
| | + | fillColor:"rgba(220,220,220,0.5)", |
| | + | strokeColor:"rgba(220,220,220,0.8)", |
| | + | highlightFill:"rgba(220,220,220,0.75)", |
| | + | highlightStroke:"rgba(220,220,220,1)", |
| | + | data: [24, 32, 6, 4], |
| | + | } |
| | + | ] |
| | + | } |
| | + | |
| | | | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/7/7d/NEFU_China_28CC959C-9C24-4F25-81A6-A7BD6F9AF770.png" style="height:119px; width:700px" /></p>
| + | var ctx_1 = document.getElementById("slide-canvas-1").getContext("2d"); |
| | + | window.myBar = new Chart(ctx_1).Bar(chart1, {responsive: true}); |
| | | | |
| − | <p> </p>
| + | }); |
| − | | + | </script> |
| − | <p> </p>
| + | </body> |
| − | | + | </div> |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/8/8a/NEFU_China_02420433-A28A-4085-AF17-0C250FF0A0ED.png" style="height:112px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/3/37/NEFU_China_6AFC9ED0-9BC4-48F2-BD1F-472ADF2A92F7.png" style="height:403px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="color:#FF0000"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/6/67/NEFU_China_52E5F2E1-0F82-4B6D-91ED-87E7215CACF5.png" style="height:150px; width:200px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 36. 1. Marker (DL2000) ; 2.3. each for one single colony; 4. Negative control;</span><span style="font-family:arial,helvetica,sans-serif"> 5</span><span style="font-family:arial,helvetica,sans-serif">. Postive control (1128bp)</span></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><img alt="" src="https://static.igem.org/mediawiki/2015/4/47/NEFU_China_BAAEBCCD-55D3-49DD-9F51-442791AEC61B.png" style="height:102px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="color:#FF0000"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/2/22/NEFU_China_2529692B-0C96-4516-AF67-92A828B120CE.png" style="height:150px; width:200px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 37. 1. Marker (DL10000) ; 2. Double enmyze digestion (4870bp,1128bp) ; 3. digest vector (4870bp) ; 4.pcr product (1128bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong><span style="font-size:28px">Plasmid construction</span></strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"> All designed fragments needed in plasmid construction were replicated using PCR . </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong><span style="font-size:18px">1. pNZ8148-lsrA</span></strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong>Insert lsrA </strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pEASY-T5 cloning vector with LsrA) with TIANprep Mini Plasmid Kit(see protocol)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/6/6d/NEFU_China_94713099-D0AB-4D58-8DA1-3E62FFA0D8B3.png" style="height:428px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/1b/NEFU_China_7A977166-D8B0-420B-9844-F0A58C8CB665.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/17/NEFU_China_39C0A08F-096B-48C1-9764-E5F5816EC6F4.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/2/20/NEFU_China_18B85C22-8EF6-4DD2-9E14-40D95C2ABE92.png" style="height:428px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> <img alt="" src="https://static.igem.org/mediawiki/2015/e/e7/NEFU_China_0ECC36ED-F12A-4C7E-A93B-FDF1212BEED6.png" style="height:225px; width:300px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 22.1. Marker (DL2000) ; 2. Postive control (1536bp) ; 3.4. each for one single colony; 5. Negative control </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/a/a3/NEFU_China_B6CBC1C6-A178-4477-99AC-F9AC10532BCD.png" style="height:103px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="font-family:arial,helvetica,sans-serif"> <span style="color:#FF0000"> <img alt="" src="https://static.igem.org/mediawiki/2015/1/1f/NEFU_China_D8A4081C-855E-4C56-BD09-F7D0AA3900F3.png" style="height:200px; width:200px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 23. 1. Marker (DL10000) ; 2. Double enmyze digestion (3167bp,1536bp) ; 3. digest vector (3167bp) ; 4.pcr product (1536bp)</span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/b/b6/NEFU_China_B5E69330-87BC-4C0D-9F33-34658ACE5A93.png" style="height:350px; width:350px" /></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><strong>2. pNZ8148-lsrB</strong></span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pEASY-T5 cloning vector with LsrB) with TIANprep Mini Plasmid Kit(see protocol)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong>Insert lsrB </strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/2/22/NEFU_China_24BACE72-9696-4E8F-996A-A39F77BCD376.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/fb/NEFU_China_CDF8CF67-9BA9-4096-BF87-D80C0AE9C68A.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/15/NEFU_China_BA1439C7-3254-4BE4-952D-BE481A8CD057.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/d/d8/NEFU_China_3434BA7A-00BA-42E4-B1E8-1A13C4ACF4EA.png" style="height:429px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="color:#FF0000"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/5/5c/NEFU_China_F40B0503-6325-44AF-8C6B-F191A326DCA3.png" style="height:250px; width:350px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 24. 1. Marker (DL2000) ; 2. Postive control (1023bp) ; 3.4.5.6.7. each for one single colony; 8. Negative control </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/d/df/NEFU_China_2C907F60-37C0-4A41-B0E1-EAE9AF7B66AB.png" style="height:81px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="color:#FF0000"><img alt="" src="https://static.igem.org/mediawiki/2015/1/1f/NEFU_China_48721673-9D75-4F41-84D3-5C6E19FC3E5C.png" style="height:200px; width:150px" /></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 25. 1. Marker (DL10000) ; 2. Double enmyze digestion (3167bp,1023bp) ; 3. digest vector (3167bp) ; 4.pcr product (1023bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/e/e9/NEFU_China_31A684E7-E52D-4BA2-B06F-AAE167675891.png" style="height:350px; width:350px" /></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><strong>3. pNZ8148-lsrC</strong></span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pEASY-T5 cloning vector with LsrC) with TIANprep Mini Plasmid Kit( see protocol)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong>Insert lsrC </strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/1a/NEFU_China_E23AD187-ABD7-4476-A15D-1CE8F6D341C3.png" style="height:379px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/5/5c/NEFU_China_FBA645D3-053B-45F5-BF6C-FB7D9CA02BA4.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/7/7e/NEFU_China_EF7A8BD4-526E-4523-9CCB-3A647ADB2CAB.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/8/83/NEFU_China_ED5282F5-23BE-44D1-AB08-BE640545F7D2.png" style="height:428px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif"> <img alt="" src="https://static.igem.org/mediawiki/2015/7/76/NEFU_China_12A38364-2AD5-4C80-98D8-6461FDE0218A.png" style="height:156px; width:165px" /></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 26. 1. Marker (DL2000) ; 2. Postive control (1044bp) ; 3.4.5.6. each for one single colony; 7. Negative control</span> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/5/55/NEFU_China_941806DA-0880-4966-B717-B339ABD5E15F.png" style="height:81px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> </span> <span style="color:#FF0000"> <span style="font-family:arial,helvetica,sans-serif">Fig 27. 1. Marker (DL10000) ; 2. Double enmyze digestion (3167bp,1044bp) ; 3. digest vector (3167bp) ; 4.pcr product (1044bp)</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/f/fb/NEFU_China_9A99958D-EDE9-4849-A907-32867121C4FF.png" style="height:350px; width:350px" /></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong><span style="font-size:18px">4. pNZ8148-lsrD</span></strong></span></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pEASY-T5 cloning vector with LsrD) with TIANprep Mini Plasmid Kit</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong>Insert lsrD </strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/7/72/NEFU_China_80F6A3BD-657A-497B-8AF4-970844664E23.png" style="height:379px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/b/b8/NEFU_China_23CDFBDF-A7D5-4660-AC6F-9AD84FB35942.png" style="height:157px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/d/da/NEFU_China_149D0F60-9342-4910-BA55-2680DC1E313B.png" style="height:119px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/4/47/NEFU_China_8794F247-02E3-4D97-8041-C9A4E7CEFDA4.png" style="height:428px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><img alt="" src="https://static.igem.org/mediawiki/2015/1/15/NEFU_China_967B6351-6F71-4272-92AA-04BF6EF14561.png" style="height:143px; width:220px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 28. 1. Marker (DL2000) ; 2. Postive control (1000bp) ; 3.4.5.6. each for one single colony; 7. Negative control </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/3/3d/NEFU_China_3D3DE586-3ED2-4F91-830A-7DCB99A41AE8.png" style="height:86px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif"> <span style="color:#FF0000"> <img alt="" src="https://static.igem.org/mediawiki/2015/9/91/NEFU_China_944D1AE5-D310-462B-8277-B98E73168B66.png" style="height:200px; width:200px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 29. 1. Marker (DL10000) ; 2. Double enmyze digestion (3167bp,1002bp) ; 3. digest vector (3167bp) ; 4.pcr product (1002bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:16px"><strong>sequencing</strong></span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/6/6c/NEFU_China_881B8214-F510-43C2-91FA-CF5AC142C518.png" style="height:350px; width:350px" /></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong><span style="font-size:18px">5. pNZ8148-lsrK</span></strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pEASY-T5 cloning vector with LsrK) with TIANprep Mini Plasmid Kit</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong>Insert lsrK</strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/3/37/NEFU_China_95C9EA81-6633-48CA-BE0F-0C0C3FAB062B.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/d/d1/NEFU_China_288A9D71-88DA-425A-967D-D25D54FBC136.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/16/NEFU_China_D51B611E-A3F0-4DBF-B64D-B108934C81A9.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/4/45/NEFU_China_C99CD053-9661-4A10-8CAE-3840AB97E4A8.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-size:24px"><span style="color:#00FFFF"><img alt="" src="https://static.igem.org/mediawiki/2015/2/28/NEFU_China_2293F963-4AF7-44BB-8BC9-1F38560C6973.png" style="height:200px; width:200px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 30. 1. Marker (DL2000) ; 2. Postive control (1593bp) ; 3.4.5. each for one single colony; 6. Negative control </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/7/7a/NEFU_China_E2E5B4B4-135E-4DA6-BD61-430962B5DDFD.png" style="height:81px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif"> <span style="color:#FF0000"> <img alt="" src="https://static.igem.org/mediawiki/2015/0/0c/NEFU_China_F7463EFC-3ABE-4F33-9ECA-32054C0D9F58.png" style="height:300px; width:300px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> </span><br />
| + | |
| − | <span style="font-family:arial,helvetica,sans-serif">Fig 31. 1. Marker (DL10000) ; 2. Double enmyze digestion (3167bp,1593bp) ; 3. digest vector (3167bp) ; 4.pcr product (1593bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/a/ad/NEFU_China_44B8EC4F-CAEA-4DCD-84EF-843DDF9AA5B8.png" style="height:350px; width:350px" /></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong><span style="font-size:18px">6. pNZ8148-lsrR</span></strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pEASY-T5 cloning vector with LsrR) with TIANprep Mini Plasmid Kit</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong>Insert lsrR</strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/a/a0/NEFU_China_96E3CC7B-17FD-4DD8-9847-242EB7BB5266.png" style="width:700px" /><span style="font-family:arial,helvetica,sans-serif"> </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/6/60/NEFU_China_C393A81F-2AC5-40D6-A4FB-F47C14B6D3E6.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/6/63/NEFU_China_D95E46ED-88DC-4C03-AAB8-8013D7CC1C3B.png" style="height:111px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/b/bf/NEFU_China_8B5B6280-B089-4BBD-B00C-3164FE9A3BF9.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="color:#FF0000"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/8/8f/NEFU_China_49AC3B23-7060-47B0-99BA-CDA1E7955793.png" style="height:123px; width:169px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 32. 1. Marker (DL2000) ; 2. Postive control (960bp) ; 3.4.5.6.7.each for one single colony; 8. Negative control </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/3/30/NEFU_China_2E868823-CAF5-46C2-982C-1B5DB3420370.png" style="height:81px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif"> <span style="color:#FF0000"><img alt="" src="https://static.igem.org/mediawiki/2015/5/58/NEFU_China_C8ED067B-D6B0-4AEB-9056-7F6286556087.png" style="height:300px; width:300px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> </span><br />
| + | |
| − | <span style="font-family:arial,helvetica,sans-serif">Fig 33. 1. Marker (DL10000) ; 2. Double enmyze digestion (3167bp,960bp) ; 3. digest vector (3167bp) ; 4.pcr product (960bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/b/b2/NEFU_China_6EF9C459-8175-4A86-A80A-D3FECB11A99A.png" style="height:350px; width:350px" /></span></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"> </p>
| + | |
| − | <p><span style="font-size:20px"><strong><span style="font-family:arial,helvetica,sans-serif">8. pBBR1MCS-5-lsrA</span></strong></span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Insert lsrA </span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pNZ8148 with LsrA) with TIANprep Mini Plasmid Kit</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-size:14px"><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/c/c2/NEFU_China_93E23CE5-9BC9-46AA-8B81-EB60A6154CBA.png" style="height:348px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/f9/NEFU_China_DBC8232D-3C4F-45A1-B117-880DD13B520E.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/e/e1/NEFU_China_34C18741-B024-438E-A27C-68B7A2495375.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/fe/NEFU_China_11E19294-D9B6-46C5-8CCD-09991F733E28.png" style="height:427px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/8/83/NEFU_China_325A7E57-B20E-4848-838D-9BFD4B801241.png" style="height:250px; width:300px" /></span></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 38. 1. Marker (DL2000) ; 2. Postive control (1952bp) ; 3.4.5.6.7. each for one single colony; 8. Negative control </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/c/cf/NEFU_China_8D7F0BEE-3055-456B-9200-6513B3EF7E05.png" style="height:81px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/8/80/NEFU_China_240B221E-02C5-4D9A-AA79-6CB566ED3546.png" style="height:170px; width:200px" /></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> </span> <span style="font-family:arial,helvetica,sans-serif"> Fig 39. 1. Marker (DL10000) ; 2. Double enmyze digestion (4707bp,1952bp) ; 3. digest vector (4707bp) ; 4.pcr product (1952bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-size:20px"><strong><span style="font-family:arial,helvetica,sans-serif">9. pBBR1MCS-5-lsrC</span></strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pNZ8148 with LsrC) with TIANprep Mini Plasmid Kit</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Insert lsrC </span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/19/NEFU_China_67956AB6-06C7-4B1D-813A-C8C06B27DB44.png" style="height:428px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/1b/NEFU_China_F2FDDE95-1E32-4CDB-9271-A6446D7B41C4.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/fc/NEFU_China_401376A0-12F4-4A49-B3D9-DD005FC17FA8.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/7/7f/NEFU_China_E94743DF-F79B-4D8E-BB03-E25EC77F9AF8.png" style="height:420px; width:700px" /></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-size:28px"><span style="color:#00FFFF"><img alt="" src="https://static.igem.org/mediawiki/2015/7/7c/NEFU_China_61F8EE29-8DAA-4232-8A60-C3F36DAECF37.png" style="height:150px; width:150px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 40. 1. Marker (DL2000) ; 2. Postive control (1503bp) ; 3.4.5.6. each for one single colony; 7. Negative control</span> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/6/64/NEFU_China_95207A9B-71D0-414F-92F3-B318A86DA925.png" style="height:81px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/b/be/NEFU_China_3089B91F-DA4F-4575-99EC-6894AA28BD32.png" style="height:300px; width:300px" /></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 41. 1. Marker (DL10000) ; 2. Double enmyze digestion (4707bp,1503bp) ; 3. digest vector (4707bp) ; 4.pcr product (1503bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-size:20px"><strong><span style="font-family:arial,helvetica,sans-serif">10. pBBR1MCS-5-lsrD</span></strong></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Miniprep (pNZ8148 with LsrD) with TIANprep Mini Plasmid Kit</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><strong>Insert lsrD</strong> </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/9/9d/NEFU_China_F478B484-1697-46AB-BE81-40B946C8C959.png" style="height:428px; width:700px" /></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Double digestion (Takara)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/2/2b/NEFU_China_705F260A-117F-48E7-B075-D788F8C18E78.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation (TaKaRa DNA Ligation Kit Ver.2.1<a href="http://www.takara.com.cn/?action=Page&Plat=pdetail&newsid=101&subclass=1">see manual</a>)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/e/eb/NEFU_China_033F7B8F-50B3-460B-89FA-1A05F4AE9F53.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation <a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/0/0a/NEFU_China_DB561A91-6641-470A-A160-6396F52823A8.png" style="height:420px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/e/ef/NEFU_China_26803078-6A16-4F32-BA69-60A8E49B49BF.png" style="height:200px; width:200px" /></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 42. 1. Marker (DL2000) ; 2.3.4.5 each for one single colony (1439bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Two-step enzyme digestion(Takara) for detection</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/7/75/NEFU_China_566EB48F-EDC1-410B-B218-AEB3CC949CCC.png" style="height:81px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="color:#FF0000"> <span style="font-size:36px"> </span></span><span style="font-size:36px"><span style="color:#00FFFF"> </span></span><img alt="" src="https://static.igem.org/mediawiki/2015/6/6a/NEFU_China_40E915B9-202B-4ACC-BC28-7A308B5A28CF.png" style="height:275px; width:250px" /><span style="font-size:36px"><span style="color:#00FFFF"> </span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 43. 1. Marker (DL10000) ; 2. </span><span style="font-family:arial,helvetica,sans-serif">digest vector (4707bp);</span><span style="font-family:arial,helvetica,sans-serif"> </span><span style="font-family:arial,helvetica,sans-serif">3. </span><span style="font-family:arial,helvetica,sans-serif">Double enmyze digestion (4707bp,1439</span><span style="font-family:arial,helvetica,sans-serif">bp) ;</span><span style="font-family:arial,helvetica,sans-serif"> 4.pcr product (139</span><span style="font-family:arial,helvetica,sans-serif">bp)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">The sequencing results are consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"> </p>
| + | |
| − | <p><strong><span style="font-size:20px"><span style="font-family:arial,helvetica,sans-serif">Transformation in <em>Lactobacillus</em></span></span></strong></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-size:18px"><span style="font-family:arial,helvetica,sans-serif">Electrotransformation of LsrBRK</span></span></strong></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Miniprep (pNZ8148–<em>LsrB</em>, pNZ8148–<em>LsrR</em>, pNZ8148–<em>LsrK</em>) with TIANprep Mini Plasmid Kit</span></strong></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">1.Preparation of competent cells </span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">100 μl bacterial culture were inoculated into 50ml of MRS and incubated at 37°C. After overnight growth, a culture at the beginning of the stationary phase was harvested by centrifugation. The bacteria were washed three times with cold electroporation buffer (PB).The cells were then resuspended in PB to an OD600 of about 50.</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">2.Electrotransfection </span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">100 μl of the cell suspension was mixed with 10 μl plasmid DNA(pNZ8148–<em>LsrB</em>, pNZ8148–<em>LsrR</em>, pNZ8148–<em>LsrK</em>). The sample was subjected to a 2.4kV, 200Ω, 25μF electric pulse in a 0.2cm cuvette.</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">3.Recovery of transformants</span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">950 μl SMRS was immediately added, and the cells were incubated for 2 h at 37°C before they were plated on MRS supplemented with the appropriate antibiotic. The plates were incubated at 37°C for 2 to 3 days under anaerobic conditions.</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">4.Clone identify</span></strong></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/e/e6/NEFU_China_2A32A9E4-41BC-4EF5-A396-74EA013FE6B9.png" style="height:592px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">We repeated this transformation process for several times. Unfortunatelly, not a single colony was grown the plate.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Then we changed the parameters of electric pulse and tried several recovery medium, but we just got some unsatisfactory results. </span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/e/ef/NEFU_China_0D70DF22-05F0-4047-AF6D-436D352FF324.png" style="height:300px; width:300px" /><span style="background-color:#EE82EE">标泳道</span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 44. 1. Marker (DL2000) ; 2.3.4.5.6.7 each for one single LsrB colony; 8.9.10.11.12.13 each for one single LsrR colony;</span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">1. Marker (DL2000) ; 2.3.4.5.6.7 each for one single LsrKcolony </span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> <img alt="" src="https://static.igem.org/mediawiki/2015/5/5c/NEFU_China_ACF45E84-42B9-4F39-87D0-EEC0B33E20E5.png" style="height:300px; width:300px" /><span style="background-color:#EE82EE">标泳道 (找我)</span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 45. 1. Marker (DL2000) ; 2.3.4.5.6.7 each for one single LsrB colony; 8.9.10.11.12.13 each for one single LsrR colony;</span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">1. Marker (DL2000) ; 2.3.4.5.6.7 each for one single LsrKcolony </span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/a/ad/NEFU_China_0C3ED3F4-8E71-4A6A-94CA-77035E009229.png" style="height:400px; width:400px" /></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 46. 1. Marker (DL10000) ; 2.3.4.5.6. each for one single LsrB colony; 7.8.9. each for one single LsrR colony;</span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> 1. Marker (DL10</span><span style="font-family:arial,helvetica,sans-serif">000) ; 2.3. each for one single LsrR colony; 4.5.6.7.</span><span style="font-family:arial,helvetica,sans-serif">8.</span><span style="font-family:arial,helvetica,sans-serif">each for one single Ls</span><span style="font-family:arial,helvetica,sans-serif">rk</span><span style="font-family:arial,helvetica,sans-serif"> colony</span><span style="font-family:arial,helvetica,sans-serif">; </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Finally, we realized it was the concentration of the plasmids which led to the low efficiency in transformation. We then did plasmid Maxiprep (see protocol) and concentration, and obtained plasmids in a concentraion of about 3000ng/μL. We also added 100μL denatured salmon sperm DNA to the cuvette before electroporation. This time, we eventually gained positive colonies.</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Clone identify</span></strong></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/c/cb/NEFU_China_1400A498-4E1E-406C-B782-5A89D481386C.png" style="height:450px; width:450px" /><span style="background-color:#EE82EE">标泳道</span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align: center;"><span style="font-family:arial,helvetica,sans-serif">Fig 47. 1. Marker (DL4000) ; 2.3.4.5.6. each for one single LsrB colony; 7. Postive control (1023bp) ; 8.9.10.11.12. each for one single colony ; 13</span><span style="font-family:arial,helvetica,sans-serif">. Postive control (1593bp)</span></p>
| + | |
| − | | + | |
| − | <p style="text-align: center;"><span style="font-family:arial,helvetica,sans-serif"> 1. Marker (DL4000) ; 2.3.4.5.6. each for one single LsrR colony; 7</span><span style="font-family:arial,helvetica,sans-serif">. Postive control (960bp);</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Electrotransformation of <em>plsr+blue </em>pigment+T</span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Maxiprep (see protocol) and concentration of pHY300PLK-pLsr- amilCP。</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Clone identify</span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/12/NEFU_China_25F1499D-4D64-42D5-8B98-EA16935BE2C3.png" style="height:420px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/0/03/NEFU_China_4E03A65D-FD94-4FCD-9E63-5EE7937F8FE5.png" style="height:200px; width:200px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 48.1. Marker (DL2000) ; 2. Postive control (1128bp) ; 3.4. each for one single colony; 5. Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"> The transformation procedures are similar to Electrotransformation of BRK</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-family:arial,helvetica,sans-serif">Clone identify</span></strong></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> </p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/7/7d/NEFU_China_5F2D24C3-D76A-46F5-9A59-C1BCAB3A2271.png" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig.?: 1. Marker (DL2000) ; 2.3.4.5.6. each for one single colony (486bp) ; 7. Postive control</span> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="color:#00FFFF">学姐需要一点描述,菌液pcr鉴定pnz9530 ,说我们设计了质粒中的一段特以引物来鉴定。</span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> </p>
| + | |
| − | <p><strong><span style="font-family:comic sans ms,cursive"><span style="font-size:28px">Labnote</span></span></strong></p>
| + | |
| − | | + | |
| − | <hr />
| + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:20px">Isolation of genomic DNA from bacteria</span></span> <span style="font-family:arial,helvetica,sans-serif">The genomic DNA of <em>Salmonella enterica</em> subsp. <em>enterica serovar Typhimurium</em> str. LT2 and<em> Lactobacillus delbrueckii</em> subsp.<em> Bulgaricus</em> str. Lb14 were isolated using the TIANamp Bacteria DNA Kit (see protocol). The concentration of genomic DNA was measured with a NanoDrop spectrophotometer. </span> </p>
| + | |
| − | | + | |
| − | <p><span style="font-size:20px"><span style="font-family:arial,helvetica,sans-serif">Strain identification</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">We applied 16S ribosomal DNA identification to identify the strain of Salmonella and Lactobacillus bought from a biotech company. Universal primers were designed according to the conserved sequences.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-size:20px"><span style="font-family:arial,helvetica,sans-serif">A. Identification of Salmonella typhimurium</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1.PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/0/05/NEFU_China_5351A137-7EAB-4153-B27D-FDEDBACC8656.png" style="height:393px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/0/08/NEFU_China_1A00253D-C888-4317-83AB-12FBD6D8F14E.png" style="height:225px; width:150px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif"> Fig 1. 1. Marker (2000bp DNA Ladder) ; 2.<span style="font-size:16px"> </span><span style="font-size:14px"><em>Salmonella typhimurium</em></span><span style="font-size:16px"> </span>16s rRNA</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">2. Ligation (TaKaRa DNA Ligation Kit see protocol)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/6/6c/NEFU_China_CD600935-F7B2-41DF-AE7A-1356B2D74459.png" style="height:117px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">3. Transformation (<a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a>)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">4. Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/fa/NEFU_China_80678D69-C533-4A86-A51D-001697EC74D8.png" style="height:380px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="color:#00FFFF"><span style="font-size:28px"><img alt="" src="https://static.igem.org/mediawiki/2015/c/ce/NEFU_China_418396AA-6D58-4C22-ADA9-5820CEC9AD9A.png" style="height:122px; width:200px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 2. 1. Marker (DL2000) ; 2.3.4.5 each for one single colony; 6. Negative control</span> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">5.Sequencing(Show results)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">6.We then did alignment using BLAST.</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/a/af/NEFU_China_E5ADCBCB-A89F-436F-AEBC-929E7CD914BD.png" style="height:461px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"> The sequence was consist with that of <em>Salmonella enterica</em> subsp. <em>enterica serovar Typhimurium</em> str. LT2.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:20px">B. Identification of Lactobacillus bulgaricus</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">1. PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/6/6d/NEFU_China_D7858136-01CC-46B3-81F6-968BA3780EF0.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"> <img alt="" src="https://static.igem.org/mediawiki/2015/b/b5/NEFU_China_3722D6C0-A70A-4DBB-B538-ED40DC3D889D.png" style="height:150px; width:100px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 3. 1. Marker (DL2000) ; 2. <span style="font-size:14px"><em>Lactobacillus bulgaricus </em>16s rRNA</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">2. Ligation (TaKaRa DNA Ligation Kit see protocol)</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/d/d6/NEFU_China_1C19B503-1B40-4A7E-A648-1BCD39B79DAC.png" style="height:117px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">3. Transformation (<a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a>)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">4. Colony PCR</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/10/NEFU_China_A09473EB-D1C4-4E0E-84C1-166D737F2C94.png" style="height:399px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p> <span style="color:#00FFFF"><span style="font-size:28px"><img alt="" src="https://static.igem.org/mediawiki/2015/6/6a/NEFU_China_087DB8ED-A25B-471E-903F-30722BDBA545.png" style="height:127px; width:200px" /></span></span></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 4. 1. Marker (DL2000) ; 2.</span><span style="font-family:arial,helvetica,sans-serif">3.4.5.6 </span><span style="font-family:arial,helvetica,sans-serif">each for one single colony; 7</span><span style="font-family:arial,helvetica,sans-serif">. Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">5. Sequencing(Show results)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">6. We then did alignment using BLAST.</span></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="font-family:arial,helvetica,sans-serif">The sequence was almost consist with that of <em>Lactobacillus delbrueckii</em> subsp.<em> Bulgaricus</em> str. Lb14. The differences were caused by subculturing perhaps.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:20px">Gene Cloning of<em> Lsr A, B, C, D, R </em>and <em>K</em></span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:16px">1. </span><span style="font-size:18px">Gene isolation using PCR</span> </span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">(Restriction enzyme sites are indicated in red font)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><em>lsrA</em></span></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/3/3c/NEFU_China_890BDFC2-9990-40D4-897D-CFA4E7E07181.png" style="height:353px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><em>lsrB</em></span></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/2/23/NEFU_China_E854482B-4DC9-40E3-8445-9FF5E79E7764.png" style="height:350px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><em><span style="font-size:18px">lsrC</span></em></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/1/1c/NEFU_China_084987BB-1EBE-415F-9049-FEFEA75EC092.png" style="height:344px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><em><span style="font-size:18px">lsrD</span></em></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/6/66/NEFU_China_B6FF9108-6231-4FE1-9032-6A754A410CC8.png" style="height:344px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><em>lsrK</em></span></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/0/0b/NEFU_China_FD688B56-317E-43BD-8B4B-39766082AAB0.png" style="height:374px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><em>LsrR</em></span></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/f/f4/NEFU_China_EE311814-A6B0-4227-ACFC-90A1272E3097.png" style="height:428px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/1/1c/NEFU_China_9D7F3095-FE86-4E6D-853C-681D7A4B603F.jpg" style="height:253px; margin-left:350px; margin-right:350px; width:400px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 5. PCR products of lsrA, B and C 1.Marker (DL2000) ; 2.lsrA (1536bp) ; 3.Negative control; 4.lsrB (1023bp) ; 5.Negative control; 6.lsrC (1089bp) ; 7.Negative control</span><img alt="" src="https://static.igem.org/mediawiki/2015/d/dd/NEFU_China_0954A0AA-5C06-49C4-A6EE-F63B36DE7E69.jpg" style="height:275px; margin-left:10px; margin-right:10px; width:400px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 6. PCR products of lsrD, R and K 1.Marker (DL2000) ; 2.lsrD (1002bp) ; 3.Negative control; 4.lsrR (960bp) ; 5.Negative control; 6.lsrK (1593bp) ; 7.Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-size:18px"><span style="font-family:arial,helvetica,sans-serif">2.Ligation (TaKaRa DNA Ligation Kit see protocol)</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><img alt="" src="https://static.igem.org/mediawiki/2015/a/a3/NEFU_China_B2E2091E-C260-4EE2-949B-3EA94A5E2D1D.png" style="height:117px; width:700px" /><img alt="" src="https://static.igem.org/mediawiki/2015/a/a3/NEFU_China_B2E2091E-C260-4EE2-949B-3EA94A5E2D1D.png" style="height:117px; width:700px" /></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">3.Transformation (<a href="https://static.igem.org/mediawiki/2014/f/f5/NEFU_China_Transformation.pdf">see protocol</a>)</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">4.Colony PCR</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">lsrA</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><img alt="" src="https://static.igem.org/mediawiki/2015/d/d8/NEFU_China_9C5A87D7-9823-4A7F-A364-2BF5A0F9577B.png" style="height:389px; width:700px" /></span></span></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/b/ba/NEFU_China_B8CBC470-E785-4DF8-8F2E-DE83D315F6EB.jpg" style="height:277px; margin-left:350px; margin-right:350px; width:400px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 7. Clony PCR results. 1. Marker (DL2000) ; 2.3.4.5 each for one single colony (1536bp) ; 6.Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">lsrB</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><img alt="" src="https://static.igem.org/mediawiki/2015/3/3f/NEFU_China_D69BDDE6-652A-41F4-8DDA-7ACC82AAF452.png" style="width:700px" /></span></span></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/5/50/NEFU_China_5818DF91-9376-471B-8B85-7967C5F0F3B7.jpg" style="height:289px; margin-left:350px; margin-right:350px; width:400px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 8. Clony PCR results. 1. Marker (DL2000) ; 2.3.4 each for one single colony (1023) ; 5.Negative control</span></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-size:18px">lsrC</span></strong></p>
| + | |
| − | | + | |
| − | <p><strong><span style="font-size:18px"><img alt="" src="https://static.igem.org/mediawiki/2015/6/6f/NEFU_China_BAC45DDC-FF3C-4A95-9076-4080D545E24D.png" style="height:428px; width:700px" /></span></strong></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/5/5e/NEFU_China_406D2C32-9C2C-479E-A080-414583438808.jpg" style="height:350px; margin-left:350px; margin-right:350px; width:350px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 9. Clony PCR results. 1. Marker (DL2000) ; 2.3.4.5 each for one single colony (1089bp) ; 6.Negative control</span></p>
| + | |
| − | | + | |
| − | <p>sequencing</p>
| + | |
| − | | + | |
| − | <p><span style="font-size:18px">lsrD</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-size:18px"><img alt="" src="https://static.igem.org/mediawiki/2015/e/eb/NEFU_China_3B850D6B-F363-4603-95DC-E802D5BB387B.png" style="width:700px" /></span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/8/86/NEFU_China_705C5ED8-2845-48E8-B4F2-574B24E8CD6B.jpg" style="height:343px; margin-left:350px; margin-right:350px; width:350px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 10. Clony PCR results. 1. Marker (DL2000) ; 2.3.4.5 each for one single colony (1002bp) ; 6.Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">lsrK</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><img alt="" src="https://static.igem.org/mediawiki/2015/7/77/NEFU_China_B80CF3CA-0BE7-4DAB-87D7-B77B87B361BA.png" style="width:700px" /></span></span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/5/59/NEFU_China_E346D693-FE67-4890-A9AB-86FAD9F8068D.jpg" style="height:350px; margin-left:350px; margin-right:350px; width:350px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 11. Clony PCR results. 1. Marker (DL2000) ; 2.3.4.5 each for one single colony(1593bp) ; 6.Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">lsrR</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px"><img alt="" src="https://static.igem.org/mediawiki/2015/c/ca/NEFU_China_4EE6A139-BDFC-4DBB-AF24-2C55019DDE48.png" style="width:700px" /></span></span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/8/83/NEFU_China_0E266A05-C035-4B64-A7F3-2A86DD198974.jpg" style="height:390px; margin-left:350px; margin-right:350px; width:350px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 12. Clony PCR results. 1. Marker (DL2000) ; 2.3.4.5 each for one single colony; 6.Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">5.sequencing</span></span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/8/85/NEFU_China_1D4FCA4D-2694-4D92-BAE4-D29C27A89915.png" style="height:370px; width:900px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 13. Sequencing result of LsrA</span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/c/c5/NEFU_China_6BD935D8-F97F-4490-81E5-6A5ECBB665FC.png" style="height:216px; width:900px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 14. Sequencing result of LsrB</span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/2/21/NEFU_China_82ED9097-982D-454B-98B5-AB52A5211EF1.png" style="height:311px; width:900px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 15. Sequencing result of LsrC</span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/d/df/NEFU_China_3C6BEB94-FDC3-410E-AEE9-676CD8312D95.png" style="height:278px; width:900px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 16. Sequencing result of LsrD</span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/8/80/NEFU_China_19EA3109-A08F-4BBD-8D6A-723289063F0E.png" style="height:415px; width:900px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 17. Sequencing result of LsrK</span></p>
| + | |
| − | | + | |
| − | <p> <img alt="" src="https://static.igem.org/mediawiki/2015/4/4f/NEFU_China_B61B8629-374E-4FC6-AF63-A2E44E7BA32F.png" style="height:277px; width:900px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 18. Sequencing result of LsrR</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">All the sequencing results were consistent with our designation, except for LsrD. There was a wrong base pair in the sequences we cloned, so we chose SOE(Splicing by overlap extension)-PCR to fix this error.</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">A. Primary PCR reaction</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Segment1-lsrD1</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/d/da/NEFU_China_8B1F2BCB-3FB8-40F7-98C8-9173416417A8.png" style="height:411px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Segment2-lsrD2</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/0/08/NEFU_China_29EAEE3C-2E1A-41D5-BBA3-7293C61B8124.png" style="height:399px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="font-family:arial,helvetica,sans-serif"> </span><img alt="" src="https://static.igem.org/mediawiki/2015/5/55/NEFU_China_D8939993-F487-49F1-B603-47E5C8A9E385.png" style="height:200px; width:200px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 19. LsrD1, LsrD2</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">B. Overlapping and elongation</span></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/3/35/NEFU_China_291C996B-3652-4073-8F85-4351A3B8BCA0.png" style="height:307px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:18px">C. Second PCR reaction</span></span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/c/c0/NEFU_China_032E37C2-7F01-4D46-8AE5-18B072CF5100.png" style="height:386px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><span style="color:#FFD700"><span style="font-family:arial,helvetica,sans-serif">图</span><span style="font-family:arial,helvetica,sans-serif">电泳</span></span></p>
| + | |
| − | | + | |
| − | <p style="margin-left:18pt; text-align:center"><span style="color:#0000FF"><span style="font-family:arial,helvetica,sans-serif">Fig 20. 1. LsrD1,2.LsrD2,3.LsrD</span></span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Ligation</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/b/bb/NEFU_China_8EF057C0-7F20-44BD-9362-D3CCBBF166BB.png" style="height:110px; width:700px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Transformation (see protocol)</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Clone pcr </span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/3/3b/NEFU_China_EB92ADF4-1C00-4F5F-BA7F-C0A4111A95C6.png" style="width:700px" /></p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><img alt="" src="https://static.igem.org/mediawiki/2015/2/23/NEFU_China_ECFDFF16-BEA0-4F4C-8EF0-D80F6D7942D4.png" style="height:150px; width:200px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="text-align:center"><span style="font-family:arial,helvetica,sans-serif">Fig 21. 1. Marker (DL2000) ; 2.3.4.5.each for one single colony(1002bp) ; 6.Negative control</span></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">Sequencing</span></p>
| + | |
| − | | + | |
| − | <p><img alt="" src="https://static.igem.org/mediawiki/2015/9/99/NEFU_China_E6C39C29-FC7F-43D2-ACD5-B1EC15673CD5.png" style="height:171px; width:552px" /></p>
| + | |
| − | | + | |
| − | <p><span style="font-family:arial,helvetica,sans-serif">As we see, the sequencing result is consistent with our designation.</span></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p style="margin-left:18.0000pt"><img alt="" src="https://static.igem.org/mediawiki/2015/5/55/NEFU_China_D8939993-F487-49F1-B603-47E5C8A9E385.png" style="height:150px; width:150px" /></p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | | + | |
| − | <p> </p>
| + | |
| − | <p><strong><span style="font-family:comic sans ms,cursive"><span style="font-size:28px">Monthly</span></span></strong></p>
| + | |
| − | | + | |
| − | <hr />
| + | |
| − | <p><br />
| + | |
| − | <span style="font-family:arial,helvetica,sans-serif"><strong><span style="font-size:18px">January</span></strong><br />
| + | |
| − | <span style="font-size:14px">Our junior and senior students were attracted by the recruiting announcement for the iGEM 2015 in our university. After several interviews and competitive selections, our team was basically created.</span><br />
| + | |
| − | <strong><span style="font-size:18px">February</span></strong><br />
| + | |
| − | <span style="font-size:14px">Our team was growing. We communicated with previous iGEMers in our school to get more insights into iGEM. Meanwhile, we started to watch successful iGEM cases of previous years.</span><br />
| + | |
| − | <strong><span style="font-size:18px">March</span></strong><br />
| + | |
| − | <span style="font-size:14px">We wandered among wikis and gradually realized what the basic factors are for a good project. Brainstorms and weekly meet-ups promoted us to discover the quintessence of iGEM.</span><br />
| + | |
| − | <strong><span style="font-size:18px">April</span></strong><br />
| + | |
| − | <span style="font-size:14px">We urged ourselves to get deeper insights into those previous cases to catch their inspiration. After meet-ups with teammates and our instructors, a draft idea of a yogurt guarder came to our mind. To make sure our project was worth proceeding, we did a survey to find out how much attention people would pay to the quality of yogurt. Fortunately, we got the expected results, although we knew we still had a long way to go.</span><br />
| + | |
| − | <strong><span style="font-size:18px">May</span></strong></span><br />
| + | |
| − | <span style="font-family:arial,helvetica,sans-serif"><span style="font-size:14px">We primarily designed the basic structure of our project. Then we read numerous papers to make an integrated protocol of our lab work.</span></span><span style="font-family:arial,helvetica,sans-serif"><span style="font-size:14px">Besides, we visited a dairy factory to learn the microbiological detection methods used in practical production of yogurt. We discovered that our method to be developed should have advantages over the traditional methods, especially in portability. </span></span><br />
| + | |
| − | <span style="font-family:arial,helvetica,sans-serif"><span style="font-size:14px">In addition, it was essential for us to adapt to the wet lab work. We started with laboratory safety training. With the instruction of our instructors and previous iGEMers, we learned a lot of techniques in molecular biology.</span><br />
| + | |
| − | <strong><span style="font-size:18px">June</span></strong><br />
| + | |
| − | <span style="font-size:14px">With efforts, our wet lab work went smoothly.</span><br />
| + | |
| − | <strong><span style="font-size:18px">July</span></strong><br />
| + | |
| − | <span style="font-size:14px">This month, we organized public activity to popularize the knowledge of yogurt production and its quality inspection, and remind people to store yogurt properly. </span></span><br />
| + | |
| − | <span style="font-family:arial,helvetica,sans-serif"><span style="font-size:14px">In addition, we did a survey to confirm the key factors which will attract customers’ attention if our yogurt guarder is developed into a real product. </span><br />
| + | |
| − | <strong><span style="font-size:18px">August</span></strong><br />
| + | |
| − | <span style="font-size:14px">On 13th-15th, we attended the summit of Central China iGEM Consortium held by Peking University. We shared our experience with other teams and really learned a lot from other teams. Here we want to express our sincere thanks to them! <br />
| + | |
| − | Meanwhile, our lab works still proceeded. We encountered difficulty in the electrotransformation of bacteria, which delayed our project but was eventually resolved after multiple attempts.</span><br />
| + | |
| − | <strong><span style="font-size:18px">September</span></strong><br />
| + | |
| − | <span style="font-size:14px">Time flies. We seized the time to make everything done and put great efforts to the wiki building and preparation for the Giant Jamboree.</span></span></p>
| + | |
| − | <p class="footer" style="color:white;margin-top:20px;text-align:center;"><span>Generated by </span><a href="https://2015.igem.org/Team:NEFU_China/Software">Flight iGEM</a>.</p>
| + | |
| − | </div>
| + | |
| − | <div class="fixl" style="width:300px">
| + | |
| − | <table width="100%" border="0" cellspacing="0" cellpadding="0">
| + | |
| − | <!--<tr> | + | |
| − | <td width="73"><a href="Safety.html"><img onmousemove="this.src='images/btla.png'" onmouseout="this.src='images/btl.png'" src="images/btl.png" width="73" height="73" /></a></td>
| + | |
| − | <td align="left" class="fixtit1">Safety</td>
| + | |
| − | </tr>--> | + | |
| − | </table>
| + | |
| − | </div> | + | |
| − | <a id="returnTop" href="javascript:;">Back to Top</a>
| + | |
| − | </body>
| + | |
| | </html> | | </html> |






 Flight iGEM requires you and your teammates to work online. We use CKEditor(a free, Open Source HTML text editor) to set up edit environment like Word or iWork.
Flight iGEM requires you and your teammates to work online. We use CKEditor(a free, Open Source HTML text editor) to set up edit environment like Word or iWork.