Difference between revisions of "Team:Paris Saclay/Measurement"
(→29th July) |
|||
| Line 5: | Line 5: | ||
</html> | </html> | ||
| + | =Interlab study= | ||
| + | Simultaneously of our projet, we participated at the InterlabStudy 2015*(lien) the purpose of this study is to collect fluorescent data from three different constructions with the collaboration of different iGEM teams which came from around the world. | ||
| + | We worked with E.coli as chassis, and the 3 constructions (J23101*+I13504*; J23106*+I13504* and J23117*+I13504*) were cloned in the reference plasmid: PSB1C3* | ||
| + | Controls used are the same constructions, without the I13504 | ||
| + | =Preparation of the constructions:= | ||
| + | All constructs used were transformed into DH5α strain. First of all, we rehydrated the different biobricks and transform them into cells. After this step we used the standard assembly protocol we inserted I13504 as suffix. The different DNA fragments containing promoters were cut with SpeI and PstI enzymes, and I13504 with SpeI and XbaI. We ligateed them using T4 DNA Ligase. | ||
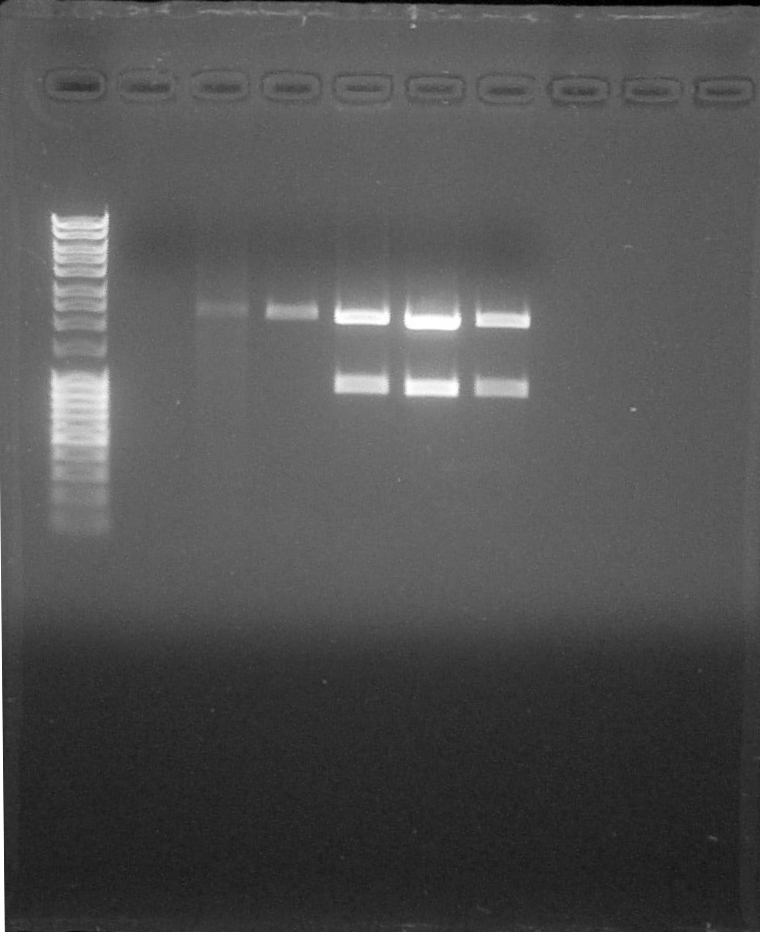
| + | *07/16 : The third last wells correspond to our 3 constructs. We observe 2 bands at 2000bp and 900bp corresponding to the expected size for the digested fragments. | ||
| + | The identity of our 3 constructs was confirmed by electrophoresis | ||
| + | |||
| + | |||
| + | =PREVIEW= | ||
| + | After taking our different colonies from the LB plate, we put them on UV lamp. The construction with J23101 had the strongest fluorescence, but the construction with J23117 shown the same fluorescence as the control. | ||
| + | |||
| + | Our bacteria were put in 5mL LB in 25mL glass tube. Kept at 37°C over night with 150rpm (position with angle in Unitron INFORS HT Incubation Shakers. | ||
| + | The morning we dilute our samples to obtain à 0,1 DO (it take one hour to pass all the control in the cytometer) | ||
| + | For the first test, it appeared that bacteria cultures were overgrown, and we observed two times the DO max so we obtain a duplicate of our sample. | ||
| + | |||
| + | =Measuring= | ||
| + | In first place we used a flux cytometer PARTEC Cyflow CUBE 6 | ||
| + | All controls correspond to the exact constructions without I13504 | ||
| + | The dark color curves correspond to the controls, the light ones tothe constructions tested. The blue one, with the other blue and the green with the other. | ||
| + | |||
| + | IMAGE | ||
| + | 3/07: the different sample were analyzed at DOmax | ||
| + | |||
| + | IMAGE | ||
| + | 30/07: We tested another biological replicate, with DO:0.6 and DOmax, The black picks are LB test. | ||
| + | |||
| + | We observe that promoter J23101 allows a 10 fold induction of fluorescence compared to J23106. Moreover J23117 has a curve response comparable to the control. | ||
| + | |||
| + | =TECAN infinite 200 Pro multimode reader= | ||
| + | Test to obtain the best excitation/emission | ||
| + | We observed with flow cytometry results that the maximum of fluorescence is obtained with J23101+GFP in stationary phase, we used these conditions to setup the experiment. | ||
| + | |||
| + | IMAGE | ||
| + | Excitation 465nm to obtain the best emission wavelength | ||
| + | |||
| + | IMAGE | ||
| + | We fixed the emission wavelength at 520nm and process to obtain the best excitation wavelength | ||
| + | |||
| + | Fluorescence Top Reading | ||
| + | Tableau | ||
| + | |||
| + | OD600#0,5 (between 0,4 and 0,55) | ||
| + | 300 µl of each sample were taken out and put on a 96-well plate (flat transparent bottom, black wall) to measure fluorescence. | ||
| + | IMAGE | ||
| + | |||
| + | We observe a pic of fluorescence with J23101+I13504, and a less one with J23106+I13504. But with the last one J23117+I13504 we observe a result like the negative control. | ||
| + | |||
| + | =Conclusion= | ||
| + | We see that the construct J23101 is the strongest of our 3 promoters. J23106 showed a lowest fold induction response, and the J23117 showed the same response as the different controls. And this result was obtain with the two different methods (flux cytometer and microplate reader) | ||
| + | When the electrophorese was done, we saw that the construct J23117+I13504 was insert in PSB1C3, but it appears that the activity of this promoter was really low or was null. | ||
| + | |||
| + | |||
=1st July= | =1st July= | ||
Rehydratation : I13504 J23117 J23106 J23101 | Rehydratation : I13504 J23117 J23106 J23101 | ||
Revision as of 17:24, 14 September 2015
Measurement
Contents
Interlab study
Simultaneously of our projet, we participated at the InterlabStudy 2015*(lien) the purpose of this study is to collect fluorescent data from three different constructions with the collaboration of different iGEM teams which came from around the world. We worked with E.coli as chassis, and the 3 constructions (J23101*+I13504*; J23106*+I13504* and J23117*+I13504*) were cloned in the reference plasmid: PSB1C3* Controls used are the same constructions, without the I13504
Preparation of the constructions:
All constructs used were transformed into DH5α strain. First of all, we rehydrated the different biobricks and transform them into cells. After this step we used the standard assembly protocol we inserted I13504 as suffix. The different DNA fragments containing promoters were cut with SpeI and PstI enzymes, and I13504 with SpeI and XbaI. We ligateed them using T4 DNA Ligase.
- 07/16 : The third last wells correspond to our 3 constructs. We observe 2 bands at 2000bp and 900bp corresponding to the expected size for the digested fragments.
The identity of our 3 constructs was confirmed by electrophoresis
PREVIEW
After taking our different colonies from the LB plate, we put them on UV lamp. The construction with J23101 had the strongest fluorescence, but the construction with J23117 shown the same fluorescence as the control.
Our bacteria were put in 5mL LB in 25mL glass tube. Kept at 37°C over night with 150rpm (position with angle in Unitron INFORS HT Incubation Shakers. The morning we dilute our samples to obtain à 0,1 DO (it take one hour to pass all the control in the cytometer) For the first test, it appeared that bacteria cultures were overgrown, and we observed two times the DO max so we obtain a duplicate of our sample.
Measuring
In first place we used a flux cytometer PARTEC Cyflow CUBE 6 All controls correspond to the exact constructions without I13504 The dark color curves correspond to the controls, the light ones tothe constructions tested. The blue one, with the other blue and the green with the other.
IMAGE 3/07: the different sample were analyzed at DOmax
IMAGE 30/07: We tested another biological replicate, with DO:0.6 and DOmax, The black picks are LB test.
We observe that promoter J23101 allows a 10 fold induction of fluorescence compared to J23106. Moreover J23117 has a curve response comparable to the control.
TECAN infinite 200 Pro multimode reader
Test to obtain the best excitation/emission We observed with flow cytometry results that the maximum of fluorescence is obtained with J23101+GFP in stationary phase, we used these conditions to setup the experiment.
IMAGE Excitation 465nm to obtain the best emission wavelength
IMAGE We fixed the emission wavelength at 520nm and process to obtain the best excitation wavelength
Fluorescence Top Reading Tableau
OD600#0,5 (between 0,4 and 0,55) 300 µl of each sample were taken out and put on a 96-well plate (flat transparent bottom, black wall) to measure fluorescence. IMAGE
We observe a pic of fluorescence with J23101+I13504, and a less one with J23106+I13504. But with the last one J23117+I13504 we observe a result like the negative control.
Conclusion
We see that the construct J23101 is the strongest of our 3 promoters. J23106 showed a lowest fold induction response, and the J23117 showed the same response as the different controls. And this result was obtain with the two different methods (flux cytometer and microplate reader) When the electrophorese was done, we saw that the construct J23117+I13504 was insert in PSB1C3, but it appears that the activity of this promoter was really low or was null.
1st July
Rehydratation : I13504 J23117 J23106 J23101
2nd July
Transformation : I13504 J23117 J23106 J23101
3rd July
Liquide culture : I13504 J23117 J23106 J23101
8th July
First Digestion:
BBa_J23101 BBa_J23106 BBa_J23117
Mix:
10µL of our plasmid with promotor 1µL SpeI 1µL PstI 2µL buffer 10x FastDigest 6µL H2O
Second Digestion:
BBa_I13504
Mix:
10µL of our plasmid with gene 1µL XbaI 1µL PstI 2µL buffer 10x FastDigest
Incubation 1h30, 37°C
9th July
Transformation:
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
On LB + Chloramphenicol 20ug/mL. Incubation ON, 37°C
15th July
Liquid culture:
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
We can observe from the plate that BBa_J23101 + BBa_I13504 and BBa_J23106 + BBa_I13504 are yellow when we expose them to the UV light. But BBa_J23117 + BBa_I13504 don't seem to be yellow on the UV light.
16th July
Digestion:
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
Reaction mix:
Plasmid: 2µL EcoRI: 0,5µL PstI: 0,5µL Buffer FastDigest (10x): 2µL H2O: 15µL
Electrophoresis:
Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET Migration 0,06A 80V
17th July
New culture on Plate
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
23rd July
Liquid culture from the 3 stocks
24th July
Cytometer
We count 500 000 events Controls:
Cells alones Cells transformed by BBa_J23101, BBa_J23106 and BBa_J23117
Our measurements: Cells transformed by
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
We uses cells in growth phase and stationary phase
Between each test, we do 2 washes with bleach and 2 washes with H2O
28th July
New culture of BBa_J23101/BBa_J23106/BBa_J23117 + BBa_I13504
Tecan utilisation:
we use only LB without chloramphenicol and we suspect a contamination of our samples. We depose in inch well 300µL We analyse the OD and fluorescence's variation (the excitation and emission wave lenght were choose after a scan in the process to obtain the best results) For each sample, we depose twelve time (12x8 plate)
- LB
- Competent cells
- Cells with J23101
- Cells with J23101 + GFP
- Cells with J23106
- Cells with J23106 + GFP
- Cells with J23117
- Cells with J23117 + GFP
We let's run for 20 cycles of 1 hour
29th July
Tecan utilisation:
This time, we use LB and chloramphenicol because the plate has just a protection and we suspect a contamination of our samples. We depose in inch well 300µL We analyse the OD and fluorescence's variation (the excitation and emission wave lenght were choose after a scan in the process to obtain the best results)
For each sample we use 3 different colonies, we depose 2 times each colonies (12x8 plate)
LB Competent cells Cells with J23101 Cells with J23101 + GFP Cells with J23106 Cells with J23106 + GFP Cells with J23117 Cells with J23117 + GFP
We let's run for 20 cycles of 1 hour
Flow cytometer
We analyse the sample see previously with ODmax and OD 0.4 But we doesn't use the good scale so we will reused it tomorrow
30th July
Flow cytometer
We count 500 000 events Controls:
LB Cells transformed by BBa_J23101, BBa_J23106 and BBa_J23117
Our measurements: Cells transformed by
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
We uses cells in growth phase and stationary phase
Between each test, we do 2 washes with bleach and 2 washes with H2O
We use a less powerful adjustment to see tall the result than the day before.