Difference between revisions of "Team:Austin UTexas/Project/Caffeine"
| Line 30: | Line 30: | ||
<b>Increase Stability</b> | <b>Increase Stability</b> | ||
<hr> | <hr> | ||
| + | <p>In addition to creating the six plasmids with combinations of genes that did not exist before</p> | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 21:49, 17 September 2015
Introduction
Background
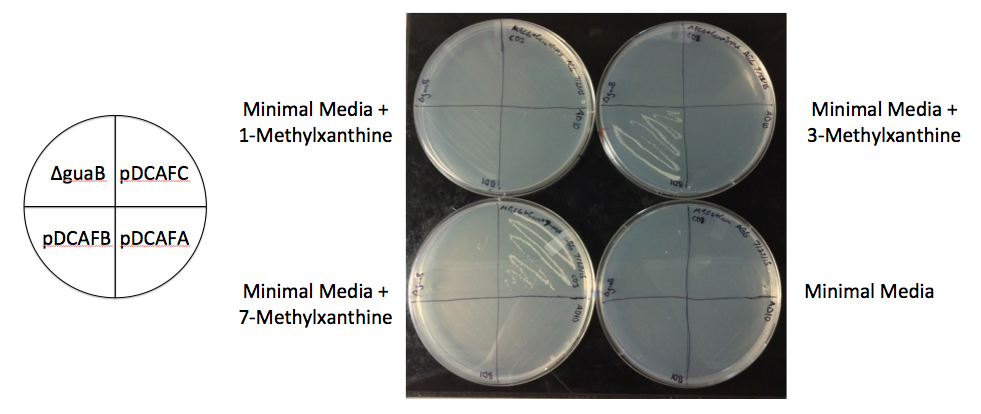
The 2012 UT Austin iGEM team created a plasmid, pDCAF3, which, when used in combination with a ∆guaB strain of E. coli, allowed them to very accurately measure the amount of caffeine in a drink. The pDCAF3 plasmid allows the E. coli to demethylate caffeine (as well as any other methylxanthine) to make xanthine. The ∆guaB strain has the guaB gene knocked out, which is a crucial step in the biosynthesis of guanine. However, if the ∆guaB strain is given the pDCAF3 plasmid, it is able to bypass the need for the enzyme encoded by guaB and make guanine. Thus, by measuring the growth of the ∆guaB E. coli with pDCAF3 plasmid in a solution containing caffeine and comparing to a growth curve, we can correctly estimate the amount of caffeine.
While this works well for solutions that only contain caffeine and no other methylxanthines, xanthine, or guanine, if any of those molecules are present, they will also contribute to the growth of the bacteria. Many of these are present in drinks made from organic products, such as coffee or tea, where knowing the exact amount of caffeine would be helpful. Thus, our goal this summer was to create a set of plasmids based on the pDCAF3 plasmid that will allow us to subtract out the background growth from those non-caffeine molecules. The pDCAF3 plasmid contains five genes that allows it to demethylate caffeine: three (ndmA, ndmB, and ndmC) that specifically demethylate each of the three methyl groups, and two (ndmD and gst9) that are necessary for the other three to work, but don’t contribute directly to the demethylation. We decided to create a set of seven plasmids that all contain ndmD and gst9, and either contain one of the other three, a pair of the other three, or all of the other three genes (Fig.1). Without all three of ndmA, ndmB, and ndmC, the bacteria will only be able to degrade some methylxanthines, not all of them, and only having all three will allow them to degrade caffeine specifically. By growing ∆guaB strains that contain each of these plasmids with a single sample of coffee, tea, or other drink that contains multiple methylxanthines, we can subtract out the growth based on the non-caffeine molecules from the strain that can degrade caffeine and get a much more accurate measurement of the amount of caffeine in the sample.
ADD THESE THINGS PLEASE:
EFM calc stuff?))
Cultures and Plates
Detection of Caffeine and Related Molecules
This set of plasmids can be used to measure the concentration of methylxanthines in solutions, even caffeinated beverages like coffee or tea that have a mixture of methylxanthines.
Caffeine and other methylxanthines
Increase Stability
In addition to creating the six plasmids with combinations of genes that did not exist before