Difference between revisions of "Team:SJTU-BioX-Shanghai/Transport Module"
(→Design of transport module) |
(→Design of transport module) |
||
| Line 9: | Line 9: | ||
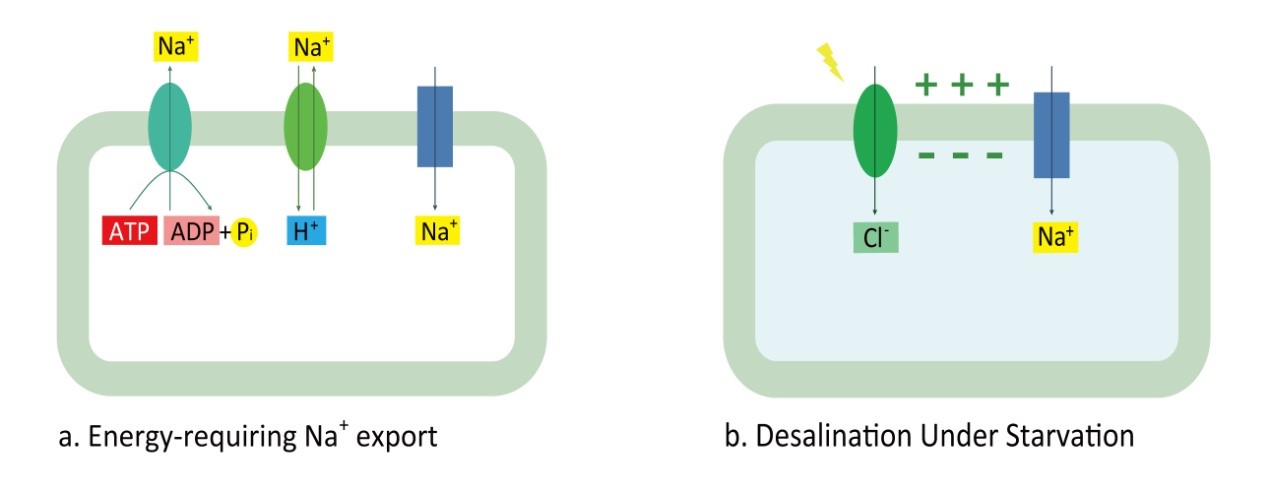
As for sodium, cyanobacteria can actively export sodium ion to maintain low internal sodium concentration under saline environment. And this is mainly performed by Na+/H+ antiporter and P-type Na+ ATPase; the former one requires H+ gradient as the energy source and the latter one consumes ATP directly. Since the ATP synthase is driven by H+ gradient, we can simply regard that ATP is required for active Na+ export. And the ATP requirement provides opportunity to halt sodium export by depleting internal ATP stores. Manipulation of cultivation conditions such as omitting photosynthetically efficient wavelengths from the light spectrum will deplete nutrient supply and exhaust ATP reserves. Moreover, it is proposed that the negative membrane potential generated by halorhodopsin would drive the influx of cation through sodium ion channels. | As for sodium, cyanobacteria can actively export sodium ion to maintain low internal sodium concentration under saline environment. And this is mainly performed by Na+/H+ antiporter and P-type Na+ ATPase; the former one requires H+ gradient as the energy source and the latter one consumes ATP directly. Since the ATP synthase is driven by H+ gradient, we can simply regard that ATP is required for active Na+ export. And the ATP requirement provides opportunity to halt sodium export by depleting internal ATP stores. Manipulation of cultivation conditions such as omitting photosynthetically efficient wavelengths from the light spectrum will deplete nutrient supply and exhaust ATP reserves. Moreover, it is proposed that the negative membrane potential generated by halorhodopsin would drive the influx of cation through sodium ion channels. | ||
| − | [[file:SJTUB_TM1.jpeg| | + | [[file:SJTUB_TM1.jpeg|700px]] |
<div style="text-indent:2em"> | <div style="text-indent:2em"> | ||
Revision as of 12:03, 18 September 2015
Design of transport module
Halorhodopsin (HR) proteins are light-driven inward-directed chloride pumps from halobacteria. They are membrane-integral proteins of the rhodopsin superfamily that form a covalent bond with the carotenoid-derived chromophore all-trans retinal. Absorption of a photon with a defined optimal wavelength induces trans-cis isomerization of retinal, which triggers a catalytic photocycle of conformational changes in the protein, resulting in the net import of one chloride per photon into the cytoplasm. (Amezaga, J. M. et al. 2014) We use it as our biodesalination driver which confers cyanobacteria the ability to absorb chloride to a significant degree.
As for sodium, cyanobacteria can actively export sodium ion to maintain low internal sodium concentration under saline environment. And this is mainly performed by Na+/H+ antiporter and P-type Na+ ATPase; the former one requires H+ gradient as the energy source and the latter one consumes ATP directly. Since the ATP synthase is driven by H+ gradient, we can simply regard that ATP is required for active Na+ export. And the ATP requirement provides opportunity to halt sodium export by depleting internal ATP stores. Manipulation of cultivation conditions such as omitting photosynthetically efficient wavelengths from the light spectrum will deplete nutrient supply and exhaust ATP reserves. Moreover, it is proposed that the negative membrane potential generated by halorhodopsin would drive the influx of cation through sodium ion channels.
Figure 1. Design of transport module. (a) Cyanobacteria actively export Na+ by Na+/H+ antiporter (green oval) and P-type Na+ ATPase (blue-green oval) to resist Na+ flowing inside through Na+ channels(Blue box) under saline environment. (b) Under starvation, ATP reserve has been exhausted and Na+ flow through Na+ channels, which is driven by the negative membrane potential generated by halorhodopsin (green oval).
In summary, the halorhodopsin drives Cl- inside, thus generating negative membrane potential, which will drive import of Na+ given that starvation inhibits active Na+ export. This design of transport module is shown in Figure 1.
We expressed halorhodopsin from in Synnechosystis sp. strain PCC 6803 with the promoters we selected, which are described in the “Selection of Promoters” section. And we did immunohistochemistry experiments to test the localization of this membrane-integral protein.
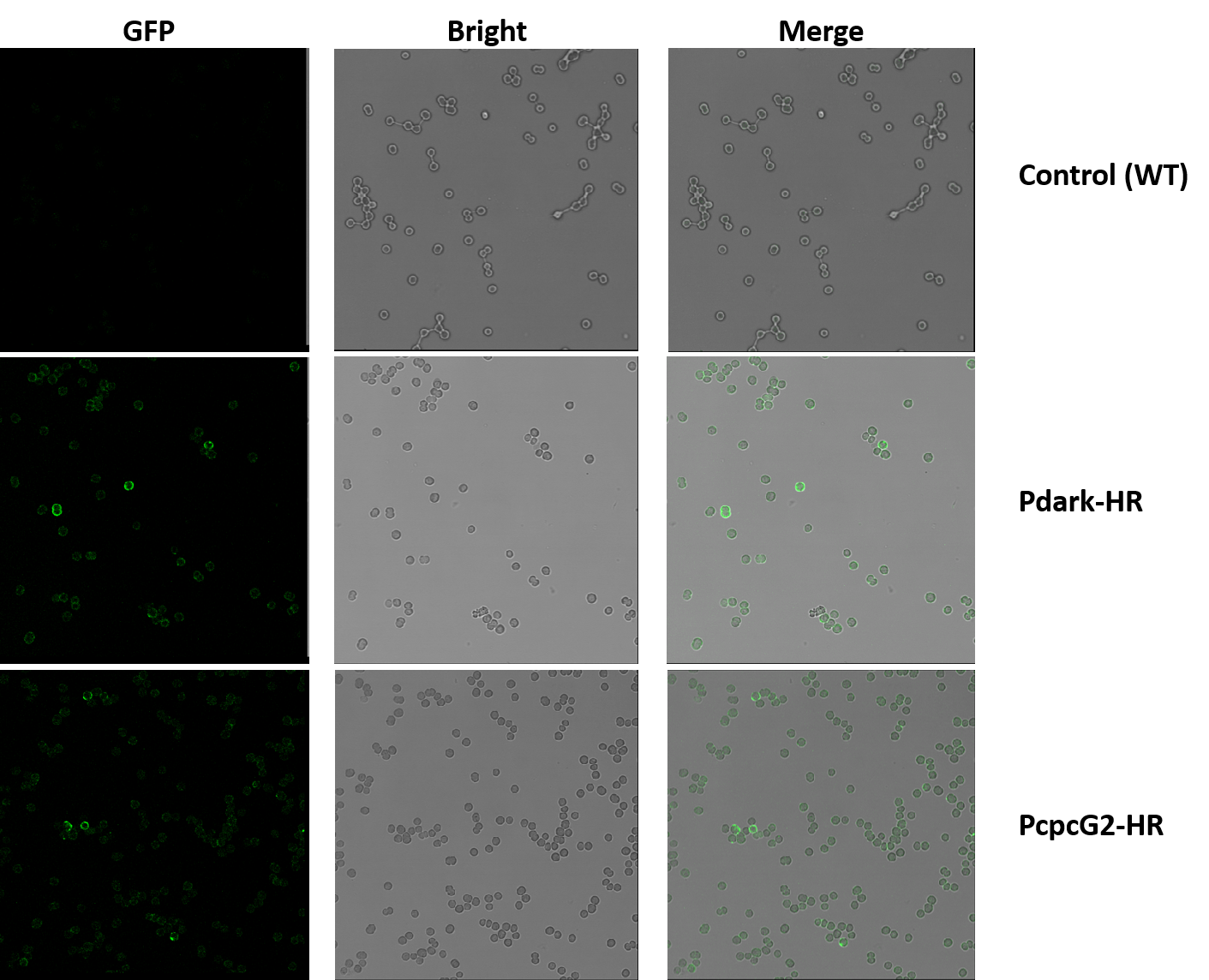
Figure 2. Detection of Membrane Localization of Halorhodopsin by Immunohistochemistry. The micrographs are shot by confocal microscope Leica SP5. Line1: control group (wildtype). Line2: experiment group1 (Pdark-HR, BBa_K1642010). Line3: experiment group2 (PcpcG2-HR, BBa_K1642011). Row1: shot under GFP field. Row2: shot under bright field. Row3: the merged micrographs.
We added His-tag to the C-terminal of the HR, thus we used anti-His mAb as the first antibody and goat anti-mouse IgG Alexa Fluor 488(green fluorescence) as the second antibody in this immunohistochemistry experiment. The pictures shot by confocal microscope is shown in Figure 2. In the experiment groups (Pdark-HR and PcpcG2-HR), many loops of green fluorescence can be seen while the control groups (wildtype) cannot, which indicates that the halorhodopsin expressed successfully on the cell membrane of the modified cyanobacteria instead of any other position in the cells.
References
Abe, K., Miyake, K., Nakamura, M., Kojima, K., Ferri, S., Ikebukuro, K., & Sode, K. (2014).Engineering of a green‐light inducible gene expression system in Synechocystis sp. PCC6803. Microbial biotechnology, 7(2), 177-183.
Amezaga, J. M., Amtmann, A., Biggs, C. A., Bond, T., Gandy, C. J., Honsbein, A., & Templeton, M. R. (2014). Biodesalination: a case study for applications of photosynthetic bacteria in water treatment. Plant physiology, 164(4), 1661-1676.
Hirose, Y., Narikawa, R., Katayama, M., & Ikeuchi, M. (2010). Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proceedings of the National Academy of Sciences, 107(19), 8854-8859.