Difference between revisions of "Team:Austin UTexas/Project/Plasmid Study"
m |
Devin wehle (Talk | contribs) |
||
| Line 143: | Line 143: | ||
The DAY 10 cultures were allowed to incubate and were sent for sequencing. A full "control" plasmid sequence was created for each culture by inserting the recorded BioBrick sequences into an existing pSB1C3-circular sequence on Benchling.com. The sequencing results were uploaded to Benchling and compared to the predicted original sequence of the plasmid for any possible mutations. | The DAY 10 cultures were allowed to incubate and were sent for sequencing. A full "control" plasmid sequence was created for each culture by inserting the recorded BioBrick sequences into an existing pSB1C3-circular sequence on Benchling.com. The sequencing results were uploaded to Benchling and compared to the predicted original sequence of the plasmid for any possible mutations. | ||
| − | |||
| − | === | + | === Results and Discussion === |
| − | + | Each plasmid created had a great amount of variance in their stability and longevity over the course of the experiment depending on the fluorescent protein that was used in the plasmid. | |
| − | + | Plasmids that bore the SYFP2 gene tended to be fairly unstable. Strains with SYFP gene tended to break down after around three days, going from very high fluorescence to no observed fluorescence. Furthermore, some samples of SYFP2 lost functionality after merely a day, showing the SYFP gene is very unstable across generations. The SYFP2 plasmids most often accumulated IS inserts, which completely removed the fluorescent protein gene from the plasmid and ceased production the protein. The most common IS elements observed were IS10R and IS10L. Furthermore, S10L/R expressed preference for a particular nucleotide location (GGCGTAGTACC). Two point mutations were also independently observed in two samples of SYFP2. | |
| − | + | Plasmids that contain the YFP gene also tended to display instability. Samples with the YFP gene often broke around five days, and a myriad of mutations appeared to have contributed to this instability. IS elements were another common feature among the YFP group. Point mutations in various locations also appeared in the YFP group, such as in the promoter and ribosome binding site. Two samples also featured a large deletion from the plasmid. | |
| − | + | The BFP group of plasmids also had moderate instability, similar to the YFP group of plasmids. The longevity of this group of plasmids was around five days before the plasmid broke down. IS elements were very common in the BFP plasmid group along with promoter deletions. | |
| − | + | ||
| − | + | Interestingly, the EYFP and ECFP plasmid groups were incredibly stable, remaining fluorescent for over ten days. Only one sample in the entire EYFP group displayed a mutation, which was a promoter deletion. Only two mutations were shown in the entire ECFP group, a large deletion and an In-del mutation. Overall, these two fluorescent proteins have shown to be very stable. See Table 1 for full results of mutations. | |
| − | + | Based on the patterns of mutations, it is clear that IS elements are a major source of instability in plasmids. Furthermore, because of the common location of insertion for IS elements, they must have a sequence preference. By changing the sequence, it is possible to increase the stability of plasmids by eliminating a mode of mutation. | |
| − | + | Furthermore, by elucidating the contribution that IS elements make towards the instability, we can better predict and reduce the instability of future plasmids. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Austin_UTexas_Footer}} | {{Austin_UTexas_Footer}} | ||
Revision as of 01:24, 19 September 2015
Measuring Fluorescent Protein Gene Stability
Introduction
One of the major issue facing genetic engineering is the overall longevity of genetic devices once inserted into organisms. An organism can be modified, but that does not ensure that the modification will last. Subsequent generations of modified organisms often lose or “break” the genetic device through mutations. Mutations that cause the inactivation of a plasmid or an inserted device decrease the metabolic load of that organism and allow the mutant and its descendants to replicate and grow faster than the originally modified organism, allowing the “broken” mutant to overtake a population.
The stability of the sequence is the focus of our research over the course of the summer. We wish to identify sequences that contribute to the relative instability of genetic devices so they can be used to predict the longevity of a genetic device as well as be taken into account when creating and modifying such devices. In the spring of 2015, E. coli was transformed using various fluorescent genes and monitored for breakage, with varying results depending on the plasmid used. Three plasmids were selected from the spring experiment to be used. Each plasmid differed by the fluorescent protein used: Yellow fluorescent protein (BBa_K592101), Super-folder Yellow fluorescent protein (BBa_K864100), and Engineered Yellow fluorescence protein (BBa_E0030).
Over the summer, these three plasmids were used to transform four strains of bacteria (TOP10, MDS42, BL21 (DE3), BW25113). Growing these transformed strains in culture and monitoring their fluorescence and stability will hopefully elucidate more regarding patterns in the sequences that contribute to the instability of genetic devices. Understanding this could lead to more stable devices and thus could increase the longevity of genetically modified organisms.
Methods
Plasmid Construction
Plasmids were constructed with standard BioBrick cloning procedures and enzymes. Composite BioBricks were constructed using the following unique BioBrick combinations:
| Plasmid | Fluorescent Protein BioBrick | Fluorescent Protein | Promoter/RBS BioBrick | Promoter;RBS Strength |
| 1 | BBa_K592100 | BFP | BBa_K608004 | Strong; Weak |
| 2 | BBa_K592100 | BFP | BBa_K608007 | Medium; Weak |
| 3 | BBa_E0020 | ECFP | BBa_K608007 | Medium; Weak |
| 4 | BBa_E0020 | ECFP | BBa_K608003 | Strong; Medium |
| 5 | BBa_E0020 | ECFP | BBa_K608002 | Strong; Strong |
| 6 | BBa_K592101 | YFP | BBa_K608002 | Strong; Strong |
| 7 | BBa_K592101 | YFP | BBa_K608006 | Medium; Medium |
| 8 | BBa_K592101 | YFP | BBa_K608007 | Medium; Weak |
| 9 | BBa_K864100 | SYFP2 | BBa_K608002 | Strong; Strong |
| 10 | BBa_K864100 | SYFP2 | BBa_K314100 | Strong; Very Strong |
| 11 | BBa_K864100 | SYFP2 | BBa_K608006 | Medium; Medium |
| 12 | BBa_E0030 | EYFP | BBa_K608007 | Medium; Weak |
| 13 | BBa_EE030 | EYFP | BBa_K608006 | Medium; Medium |
| 14 | BBa_E0030 | EYFP | BBa_K314100 | Strong; Very Strong |
Several fluorescent protein/promoter+RBS combinations were studied in more than one experiment, which is why each plasmid above is classified as a 'unique' combination.
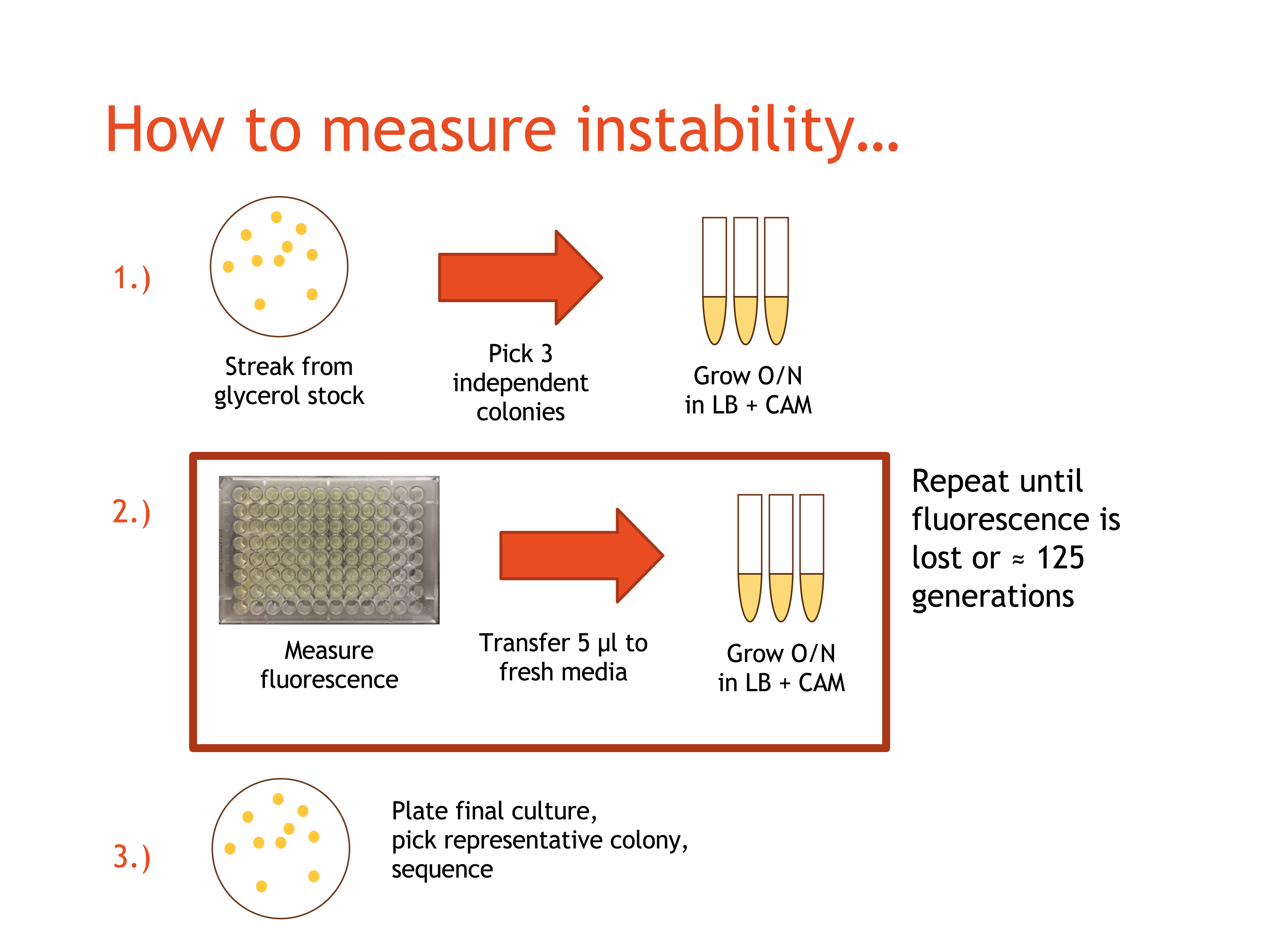
The BioBricks in each pair were ligated together using T4 DNA Ligase, and transformed into TOP10 E. coli. Prepared cultures were streaked onto LB/chloramphenicol (CAM) agar plates, and were placed in a 37°C incubator overnight. After at least 20 hours, the plates were retrieved, and from each plate three independent, fluorescent colonies were selected and each placed into 5mL of LB/CAM media. The tubes for each were labelled with the date, the culture name, DAY 1, and the strain used. The tubes were placed in a 37°C shaker-incubator and the plates were placed in a 4°C cold room for storage.
Fluorescence Readings
The primary aim of this project was to assess the fluorescent protein coding sequences that are most prone to breaking/mutating. In order to do this, we had to propagate different fluorescent proteins and record their fluorescence daily.
The day immediately following the start of the DAY 1 cultures, the DAY 1 cultures were retrieved and resuspendeded. Three empty culture tubes for each participating team member were collected and labelled the same way as the first set of culture tubes, but with DAY 2 instead of DAY 1. 5mL of LB/CAM media were placed into each culture tube, and 5μL of culture from the microcentrifuge tubes were pipetted into the appropriate culture tube. The culture tubes for DAY 2 were placed into the shaker-incubator, and the DAY 1 culture tubes were placed in the cold room.
200μL of each DAY 1 culture were pipetted into a well on a 96-well plate (which was stored on ice when not in use). The well number, date, and culture name were recorded, and the plates were read for fluorescence the following morning. This process of moving cultures forward and obtaining fluorescence readings was repeated daily until the culture fluorescence dropped dramatically, or until DAY 10, so that each culture had a maximum 10 culture tubes over the 10 days.
The DAY 10 cultures were allowed to incubate and were sent for sequencing. A full "control" plasmid sequence was created for each culture by inserting the recorded BioBrick sequences into an existing pSB1C3-circular sequence on Benchling.com. The sequencing results were uploaded to Benchling and compared to the predicted original sequence of the plasmid for any possible mutations.
Results and Discussion
Each plasmid created had a great amount of variance in their stability and longevity over the course of the experiment depending on the fluorescent protein that was used in the plasmid.
Plasmids that bore the SYFP2 gene tended to be fairly unstable. Strains with SYFP gene tended to break down after around three days, going from very high fluorescence to no observed fluorescence. Furthermore, some samples of SYFP2 lost functionality after merely a day, showing the SYFP gene is very unstable across generations. The SYFP2 plasmids most often accumulated IS inserts, which completely removed the fluorescent protein gene from the plasmid and ceased production the protein. The most common IS elements observed were IS10R and IS10L. Furthermore, S10L/R expressed preference for a particular nucleotide location (GGCGTAGTACC). Two point mutations were also independently observed in two samples of SYFP2.
Plasmids that contain the YFP gene also tended to display instability. Samples with the YFP gene often broke around five days, and a myriad of mutations appeared to have contributed to this instability. IS elements were another common feature among the YFP group. Point mutations in various locations also appeared in the YFP group, such as in the promoter and ribosome binding site. Two samples also featured a large deletion from the plasmid.
The BFP group of plasmids also had moderate instability, similar to the YFP group of plasmids. The longevity of this group of plasmids was around five days before the plasmid broke down. IS elements were very common in the BFP plasmid group along with promoter deletions.
Interestingly, the EYFP and ECFP plasmid groups were incredibly stable, remaining fluorescent for over ten days. Only one sample in the entire EYFP group displayed a mutation, which was a promoter deletion. Only two mutations were shown in the entire ECFP group, a large deletion and an In-del mutation. Overall, these two fluorescent proteins have shown to be very stable. See Table 1 for full results of mutations.
Based on the patterns of mutations, it is clear that IS elements are a major source of instability in plasmids. Furthermore, because of the common location of insertion for IS elements, they must have a sequence preference. By changing the sequence, it is possible to increase the stability of plasmids by eliminating a mode of mutation. Furthermore, by elucidating the contribution that IS elements make towards the instability, we can better predict and reduce the instability of future plasmids.