Difference between revisions of "Team:USTC-Software/Description"
Zh19970205 (Talk | contribs) |
EmmyNoether (Talk | contribs) |
||
| Line 38: | Line 38: | ||
<div id="Features"> | <div id="Features"> | ||
<h1>Features</h1> | <h1>Features</h1> | ||
| − | < | + | <ul> |
| − | <li><h3>Logic: From a truth table to abstract logic circuits</h3> | + | <li><h3>1.Logic: From a truth table to abstract logic circuits</h3> |
| − | <p>Function: </p> | + | <p><b>Function: </b></p> |
<p>In BioBLESS, a truth table dialog box will be displayed for users to input Boolean numbers 0/1. All you need to do is to click Boolean buttons and then ¡°Create¡±. The corresponding designed logic circuits with different predicted performances will be automatically generated and displayed in order. You can immediately and clearly see the configuration of a designed logic circuit, which in most cases is supposed to be a complicated multi-input circuit. Besides the function above, gates list will provide many Boolean gates, and users can drag them freely to our canvas to design logic circuits.</p> | <p>In BioBLESS, a truth table dialog box will be displayed for users to input Boolean numbers 0/1. All you need to do is to click Boolean buttons and then ¡°Create¡±. The corresponding designed logic circuits with different predicted performances will be automatically generated and displayed in order. You can immediately and clearly see the configuration of a designed logic circuit, which in most cases is supposed to be a complicated multi-input circuit. Besides the function above, gates list will provide many Boolean gates, and users can drag them freely to our canvas to design logic circuits.</p> | ||
<img width=100% id="Features1" src="/wiki/images/6/62/USTC-Software-Logic.png"/> | <img width=100% id="Features1" src="/wiki/images/6/62/USTC-Software-Logic.png"/> | ||
| − | <p>Details: </p> | + | <p><b>Details: </b></p> |
<p>One ultimate goal of synthetic biology is to design novel circuits with the aid of computers. </p> | <p>One ultimate goal of synthetic biology is to design novel circuits with the aid of computers. </p> | ||
<p>Concepts and algorithms from electrical engineering can be exploited to establish a framework for computational automatic design of gene Boolean gates and devices. Gene digital circuits can be automatically designed in an electronic fashion. Although the mechanisms underlying biological process differ in many aspects from those in electrical engineering, the framework determining the relationship between the inputs and outputs are universal only if Boolean gates constitutes the basic building blocks. </p> | <p>Concepts and algorithms from electrical engineering can be exploited to establish a framework for computational automatic design of gene Boolean gates and devices. Gene digital circuits can be automatically designed in an electronic fashion. Although the mechanisms underlying biological process differ in many aspects from those in electrical engineering, the framework determining the relationship between the inputs and outputs are universal only if Boolean gates constitutes the basic building blocks. </p> | ||
| Line 50: | Line 50: | ||
| − | <li><h3>Gene network: See the detailed structure of a designed gene circuit</h3> | + | <li><h3>2.Gene network: See the detailed structure of a designed gene circuit</h3> |
| − | <p>Function: </p> | + | <p><b>Function: </b></p> |
<p>A simple click on "Gene network" button will lead you to another tab where the detailed structure of your designed gene circuit is shown. When a gene circuit consists of many Boolean gates, it is crucial to understand the internal substances and mechanisms. In this tab, you can see how substances react with each other clearly, such as inhibition and activation in or between Boolean gates.</p> | <p>A simple click on "Gene network" button will lead you to another tab where the detailed structure of your designed gene circuit is shown. When a gene circuit consists of many Boolean gates, it is crucial to understand the internal substances and mechanisms. In this tab, you can see how substances react with each other clearly, such as inhibition and activation in or between Boolean gates.</p> | ||
<img width=100% id="Features2" src="/wiki/images/8/8a/USTC-Software-Gene_NW.png"/> | <img width=100% id="Features2" src="/wiki/images/8/8a/USTC-Software-Gene_NW.png"/> | ||
| − | <p>Details: </p> | + | <p><b>Details: </b></p> |
<p>As the scale of gene circuits grows larger, the difficulty of construction and measurement can be extremely formidable. Under the circumstances, it cannot be emphasized more to understand the internal substances and mechanisms. When the Boolean gates are combined together, the performance of the integrated system may not be as good as expected, even if the property of each Boolean gate is well understood. In contrast to the design of modern engineering disciplines which are always predictable and have a solid basis of accurate theories, synthetic biology may fall into the consistent process of trial-and-error. Huge cost of repeated experiments therefore becomes one of the constraints for biologists to further standardize Biobricks and devices. Therefore, our software is elaborately designed to provide a platform where biologists can design large-scale gene circuit before realizing them in laboratories.</p> </li> | <p>As the scale of gene circuits grows larger, the difficulty of construction and measurement can be extremely formidable. Under the circumstances, it cannot be emphasized more to understand the internal substances and mechanisms. When the Boolean gates are combined together, the performance of the integrated system may not be as good as expected, even if the property of each Boolean gate is well understood. In contrast to the design of modern engineering disciplines which are always predictable and have a solid basis of accurate theories, synthetic biology may fall into the consistent process of trial-and-error. Huge cost of repeated experiments therefore becomes one of the constraints for biologists to further standardize Biobricks and devices. Therefore, our software is elaborately designed to provide a platform where biologists can design large-scale gene circuit before realizing them in laboratories.</p> </li> | ||
| − | <li><h3>Simulation: Get concentration-time simulation results</h3> | + | <li><h3>3.Simulation: Get concentration-time simulation results</h3> |
| − | <p>Function: </p> | + | <p><b>Function: </b></p> |
<p>Given a designed gene circuit, such as a simple serial circuit composed of an AND gate and an OR gate, you can use our default parameters and change initial concentration of reactants to get simulation results. </p> | <p>Given a designed gene circuit, such as a simple serial circuit composed of an AND gate and an OR gate, you can use our default parameters and change initial concentration of reactants to get simulation results. </p> | ||
<p>If you want to get different results, you just need to change system parameters such as the rate of reactions, namely, to fill constants into the dialog boxes on our canvas. </p> | <p>If you want to get different results, you just need to change system parameters such as the rate of reactions, namely, to fill constants into the dialog boxes on our canvas. </p> | ||
| − | <p> | + | <p>What's more, simulation time slot is necessary as well to give concentration-time figures of the inputs and outputs of a given gene circuit. You are free to see the concentration-time evolution of every substance in designed gene circuits individually.</p> |
<img width=100% id="Features3" src="/wiki/images/e/e2/USTC-Software-Simulation.png"/> | <img width=100% id="Features3" src="/wiki/images/e/e2/USTC-Software-Simulation.png"/> | ||
| − | <p>Details: </p> | + | <p><b>Details: </b></p> |
<p>There are two formalisms for mathematically describing the time behavior of a spatially homogeneous chemical system.</p> | <p>There are two formalisms for mathematically describing the time behavior of a spatially homogeneous chemical system.</p> | ||
<p>The first one is the deterministic approach which regards the time evolution as a continuous and wholly predictable process governed by a set of coupled ordinary differential equations (the ¡°reaction-rate equations¡±).</p> | <p>The first one is the deterministic approach which regards the time evolution as a continuous and wholly predictable process governed by a set of coupled ordinary differential equations (the ¡°reaction-rate equations¡±).</p> | ||
| Line 75: | Line 75: | ||
<p>There is, however, a way to make exact numerical calculations within the framework of the stochastic formulation without having to deal with the master equation directly. It is a relatively simple digital computer algorithm, which uses a rigorously derived Monte Carlo procedure to numerically simulate the time evolution of a given chemical system. Like the master equation, this ¡°stochastic simulation algorithm¡± can correctly explain the inherent fluctuations and correlations that are unfortunately ignored in the deterministic formulation. </p></li> | <p>There is, however, a way to make exact numerical calculations within the framework of the stochastic formulation without having to deal with the master equation directly. It is a relatively simple digital computer algorithm, which uses a rigorously derived Monte Carlo procedure to numerically simulate the time evolution of a given chemical system. Like the master equation, this ¡°stochastic simulation algorithm¡± can correctly explain the inherent fluctuations and correlations that are unfortunately ignored in the deterministic formulation. </p></li> | ||
| − | <li><h3>Analysis: Get robustness and sensitivity analysis</h3> | + | <li><h3>4.Analysis: Get robustness and sensitivity analysis</h3> |
<td class="Featureword"> | <td class="Featureword"> | ||
| − | <p>Function: </p> | + | <p><b>Function: </b></p> |
<p>While offering various functions, BioBLESS also serves as an analysis tool for designed gene circuits. Having designed a gene circuit, you can click the ¡°Analysis¡± button to reset parameters of initial concentrations and reaction rates. Meanwhile, BioBLESS will set simulation slots and carry the simulation task automatically. According to the perturbation ratio you input, such as 20% or 30%, the concentration-time curves of inputs and outputs will be shown together to shed light on the robustness of a designed gene circuit. Moreover, data processing and demonstration will help users to better understand the robustness of designed gene circuits.</p> | <p>While offering various functions, BioBLESS also serves as an analysis tool for designed gene circuits. Having designed a gene circuit, you can click the ¡°Analysis¡± button to reset parameters of initial concentrations and reaction rates. Meanwhile, BioBLESS will set simulation slots and carry the simulation task automatically. According to the perturbation ratio you input, such as 20% or 30%, the concentration-time curves of inputs and outputs will be shown together to shed light on the robustness of a designed gene circuit. Moreover, data processing and demonstration will help users to better understand the robustness of designed gene circuits.</p> | ||
<img width=100% id="Features4" src="/wiki/images/d/df/USTC-Software-Analysis.png"/> | <img width=100% id="Features4" src="/wiki/images/d/df/USTC-Software-Analysis.png"/> | ||
| − | <p>Details: </p> | + | <p><b>Details: </b></p> |
<p>Robustness and sensitivity can measure the property of being strong and healthy or not in constitution. They refer to the ability of tolerating perturbations that might affect the system¡¯s functional body. When perturbations to the inputs of a system are small in magnitude, we hope to measure whether the concentration of outputs will change sharply or in extreme cases, even destroy the input-output logic embodied in truth tables. The most intractable problem we confront is the overload on our server to complete the simulation task. After literature reviewing, we choose some factors, for example, translation rate, to which a designed gene circuit is most sensitive. We also set a range for parameter fluctuations. Some designed circuits are proven to be robust and resistant to environment perturbations, while some other plausible well-functioning gene circuits are sensitive to specific parameters. The analysis is crucial because it gives us deeper understanding of a complicated gene circuit and lays foundation for wet-lab realization of functional novel gene circuits.</p> | <p>Robustness and sensitivity can measure the property of being strong and healthy or not in constitution. They refer to the ability of tolerating perturbations that might affect the system¡¯s functional body. When perturbations to the inputs of a system are small in magnitude, we hope to measure whether the concentration of outputs will change sharply or in extreme cases, even destroy the input-output logic embodied in truth tables. The most intractable problem we confront is the overload on our server to complete the simulation task. After literature reviewing, we choose some factors, for example, translation rate, to which a designed gene circuit is most sensitive. We also set a range for parameter fluctuations. Some designed circuits are proven to be robust and resistant to environment perturbations, while some other plausible well-functioning gene circuits are sensitive to specific parameters. The analysis is crucial because it gives us deeper understanding of a complicated gene circuit and lays foundation for wet-lab realization of functional novel gene circuits.</p> | ||
</li> | </li> | ||
| − | <li><h3>DNA: Get the DNA sequence</h3> | + | <li><h3>5.DNA: Get the DNA sequence</h3> |
| − | <p>Function: </p> | + | <p><b>Function: </b></p> |
<p>After designing gene circuits, the DNA sequence and other information of the designed circuit are available for users to check and download. </p> | <p>After designing gene circuits, the DNA sequence and other information of the designed circuit are available for users to check and download. </p> | ||
<img width=100% id="Features5" src="/wiki/images/3/33/USTC-Software-DNA.png"/> | <img width=100% id="Features5" src="/wiki/images/3/33/USTC-Software-DNA.png"/> | ||
</li> | </li> | ||
| − | </ | + | </ul> |
Revision as of 01:29, 19 September 2015
- Home
- Project
- Description
- Demo
- Future Work
- Validation
- Team
- Technology
- Notebook
- Requirements
- Human Practices
- Attributions

Overview
Aiming to rationally design synthetic gene circuits with specific functions, we develop a software for automatic design which integrate methods including electrical engineering, stochastic simulation, and circuit performance analysis.
- Given a truth table where the inputs and outputs take only 0/1 values by applying the standard procedure of electrical engineering to biology, we convert the truth table into Boolean Formula and use relevant algorithm to find corresponding digital circuit scheme which is as simple as possible in the sense of electrical engineering.
- Appropriate biological structure of logic gates will be chosen from our database and embedded into the digital circuit. Among all these possible gene circuit designs, our software can recommend one or more most reliable circuits according to gene circuits’ fitness score which evaluates their complexity. Users can either accept the recommendation or choose any gene circuit they like. Meanwhile, the detailed biological structure will be screened for users’ reference.
- Once a gene circuit is generated, our software will numerically simulate the stochastic time evolution of all the inputs and outputs.
- If the simulation results correspond with the initial truth table, sensitivity analysis and optimization will be done to further maximize circuits’ practical realizablity, and robustness analysis will access their relative immunity to perturbations.
- Finally, a thoroughly considered gene circuit will be generated by our software, available for wet-lab experiments.
Our software keeps the compatibility of various methods or algorithms and reserves the space for users to redesign. Moreover, our software adopts user-friendly GUI and visualize the designing process, which makes the operations much more convenient for users.
Motivation
Since its inception over a decade ago, a series of remarkable progress have been made in the field of Synthetic Biology. As the design and testing cycles rely more on DNA synthesis and high-throughput assembly methods than on the traditional molecular cloning tools, workflow for a biological circuit engineer may no longer be limited by the pace of fabrication but instead by their ability to analyze circuit behavior in the near future. Therefore, our team adopts a workflow for biological circuit design with multiple-inputs-single-output and given functions in Boolean value. By discipline borrowed from electrical engineering and the analysis of connecting logic gates, our software is able to provide several more complex and novel circuit designs.
Meanwhile, circuit¡¯s practical realizability is very vital in order to avoid unnecessary cost and effort in wet-lab. We implement stochastic time evolution modelling to profoundly predict the behavior of circuits. The elaborated S-score, robustness and sensitivity analysis are used in evaluating and screening the circuits.
Through a well-rounded consideration of present difficulties in circuit design, we are motivated to develop such a highly integrated software that can automatically design circuits with both complexity and reliable behavior.
Features
1.Logic: From a truth table to abstract logic circuits
Function:
In BioBLESS, a truth table dialog box will be displayed for users to input Boolean numbers 0/1. All you need to do is to click Boolean buttons and then ¡°Create¡±. The corresponding designed logic circuits with different predicted performances will be automatically generated and displayed in order. You can immediately and clearly see the configuration of a designed logic circuit, which in most cases is supposed to be a complicated multi-input circuit. Besides the function above, gates list will provide many Boolean gates, and users can drag them freely to our canvas to design logic circuits.
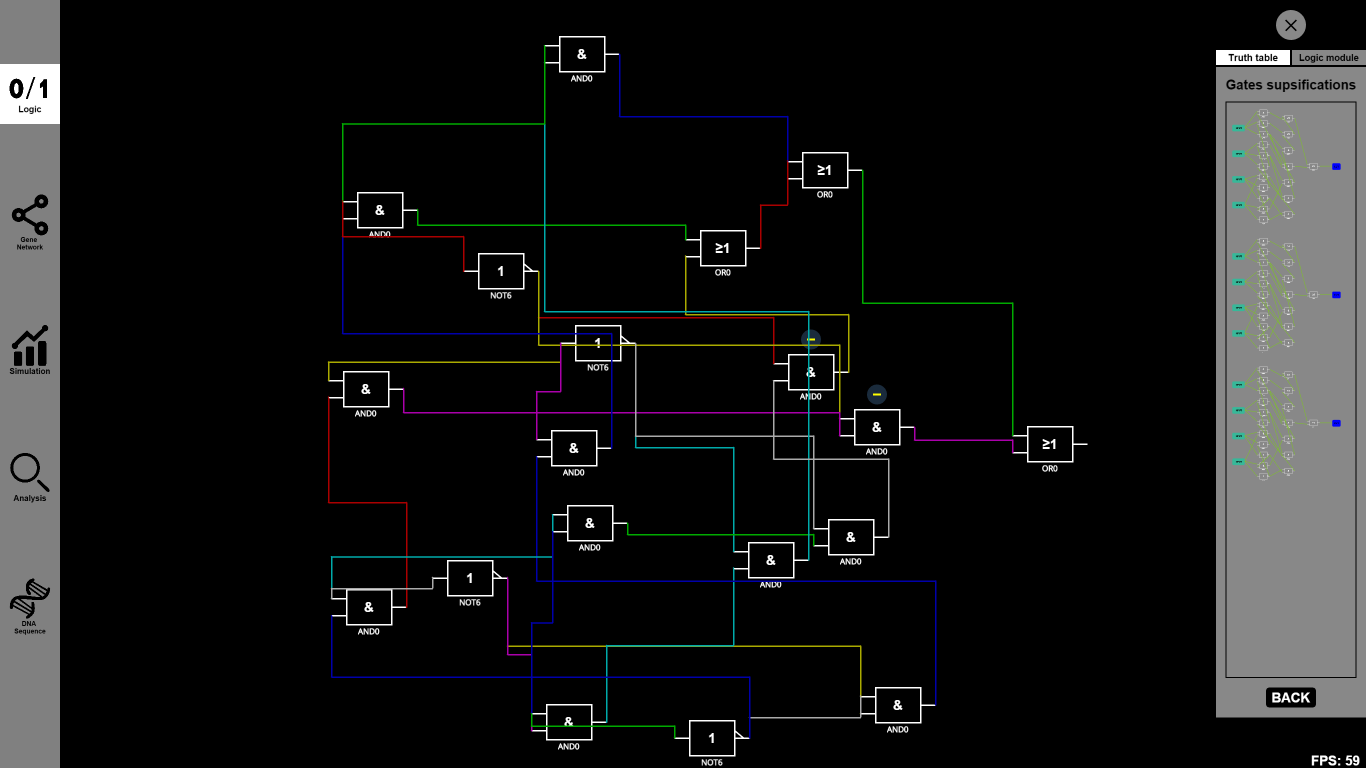
Details:
One ultimate goal of synthetic biology is to design novel circuits with the aid of computers.
Concepts and algorithms from electrical engineering can be exploited to establish a framework for computational automatic design of gene Boolean gates and devices. Gene digital circuits can be automatically designed in an electronic fashion. Although the mechanisms underlying biological process differ in many aspects from those in electrical engineering, the framework determining the relationship between the inputs and outputs are universal only if Boolean gates constitutes the basic building blocks.
Therefore with the introduction of electrical engineering, if biologists need to design some gene circuits with specific functions, they just need to input truth tables and then by employing Espresso algorithms, our software will provide a logic circuit composed of different Boolean gates and function parts. Our library of well-characterized Boolean gates is the basis for the assembly of more complicated gene digital circuits. What is equivalently crucial is that the rational design based on digital circuits will bring about the freedom of designing complicated logic relationships between inputs and outputs, but will no longer be confined to mere single-input-single-output design. In our software, eight inputs are available for users. However, it is worth mentioning that the maximum number of inputs is actually not limited to our algorithm but to the space of the user interface.
It is possible that different schemes may correspond to one expected genetic function. In order to pick out the most feasible circuit design, we weighed many contributing factors to relieve the wet-lab cells of heavy burden and tried our best to eliminate the chances of cross talk and coupling. Fitness score is introduced into our software to measure the applicability and functionality of designed gene circuits. Many synthetic biologists have introduced similar scores to rank functional gene circuits. Our fitness score combines several types of scores to give a comprehensive and reasonable evaluation. Different scores put different emphasis on genetic regulatory processes which can also be influenced by different experimental environments. As a result, using a set of scoring standards, rather than only single one of them, to evaluate different properties of a gene circuit is undoubtedly a better approach to ranking gene circuits.
2.Gene network: See the detailed structure of a designed gene circuit
Function:
A simple click on "Gene network" button will lead you to another tab where the detailed structure of your designed gene circuit is shown. When a gene circuit consists of many Boolean gates, it is crucial to understand the internal substances and mechanisms. In this tab, you can see how substances react with each other clearly, such as inhibition and activation in or between Boolean gates.
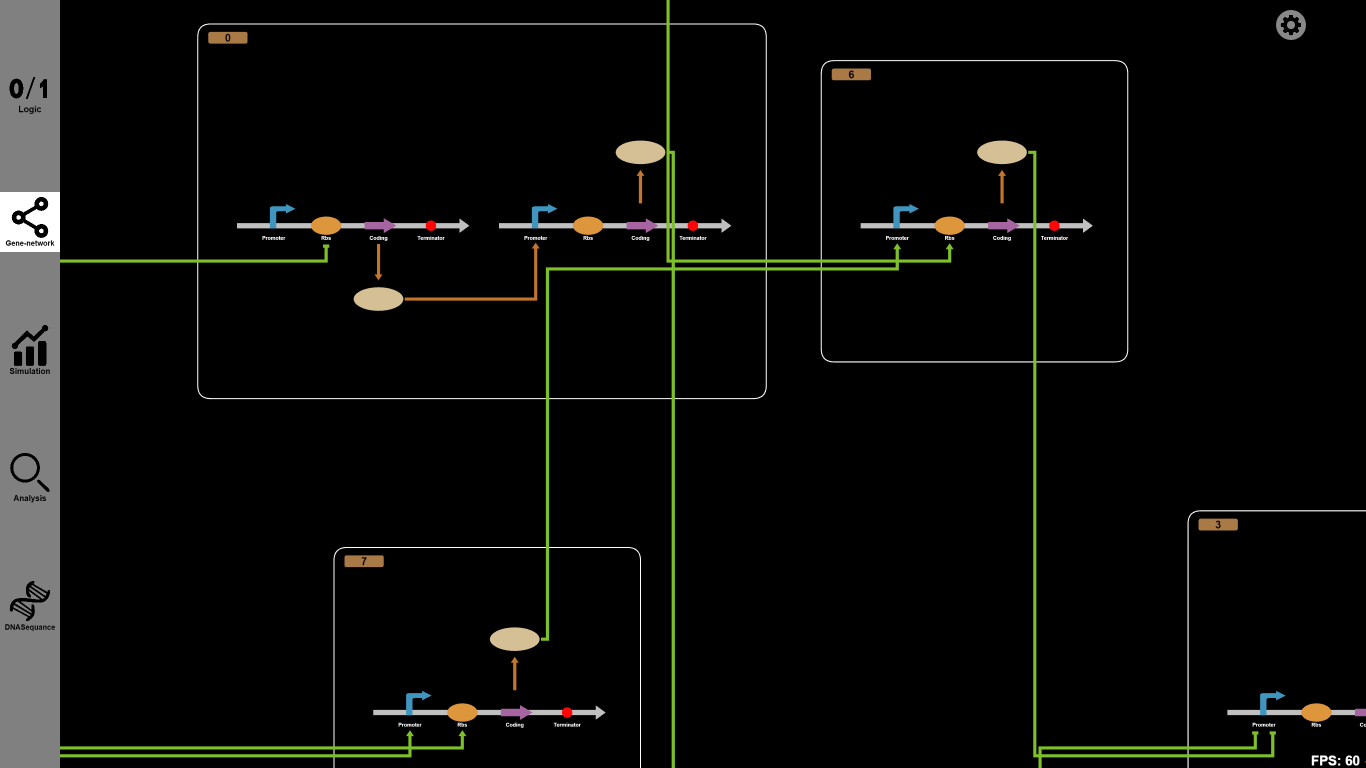
Details:
As the scale of gene circuits grows larger, the difficulty of construction and measurement can be extremely formidable. Under the circumstances, it cannot be emphasized more to understand the internal substances and mechanisms. When the Boolean gates are combined together, the performance of the integrated system may not be as good as expected, even if the property of each Boolean gate is well understood. In contrast to the design of modern engineering disciplines which are always predictable and have a solid basis of accurate theories, synthetic biology may fall into the consistent process of trial-and-error. Huge cost of repeated experiments therefore becomes one of the constraints for biologists to further standardize Biobricks and devices. Therefore, our software is elaborately designed to provide a platform where biologists can design large-scale gene circuit before realizing them in laboratories.
3.Simulation: Get concentration-time simulation results
Function:
Given a designed gene circuit, such as a simple serial circuit composed of an AND gate and an OR gate, you can use our default parameters and change initial concentration of reactants to get simulation results.
If you want to get different results, you just need to change system parameters such as the rate of reactions, namely, to fill constants into the dialog boxes on our canvas.
What's more, simulation time slot is necessary as well to give concentration-time figures of the inputs and outputs of a given gene circuit. You are free to see the concentration-time evolution of every substance in designed gene circuits individually.
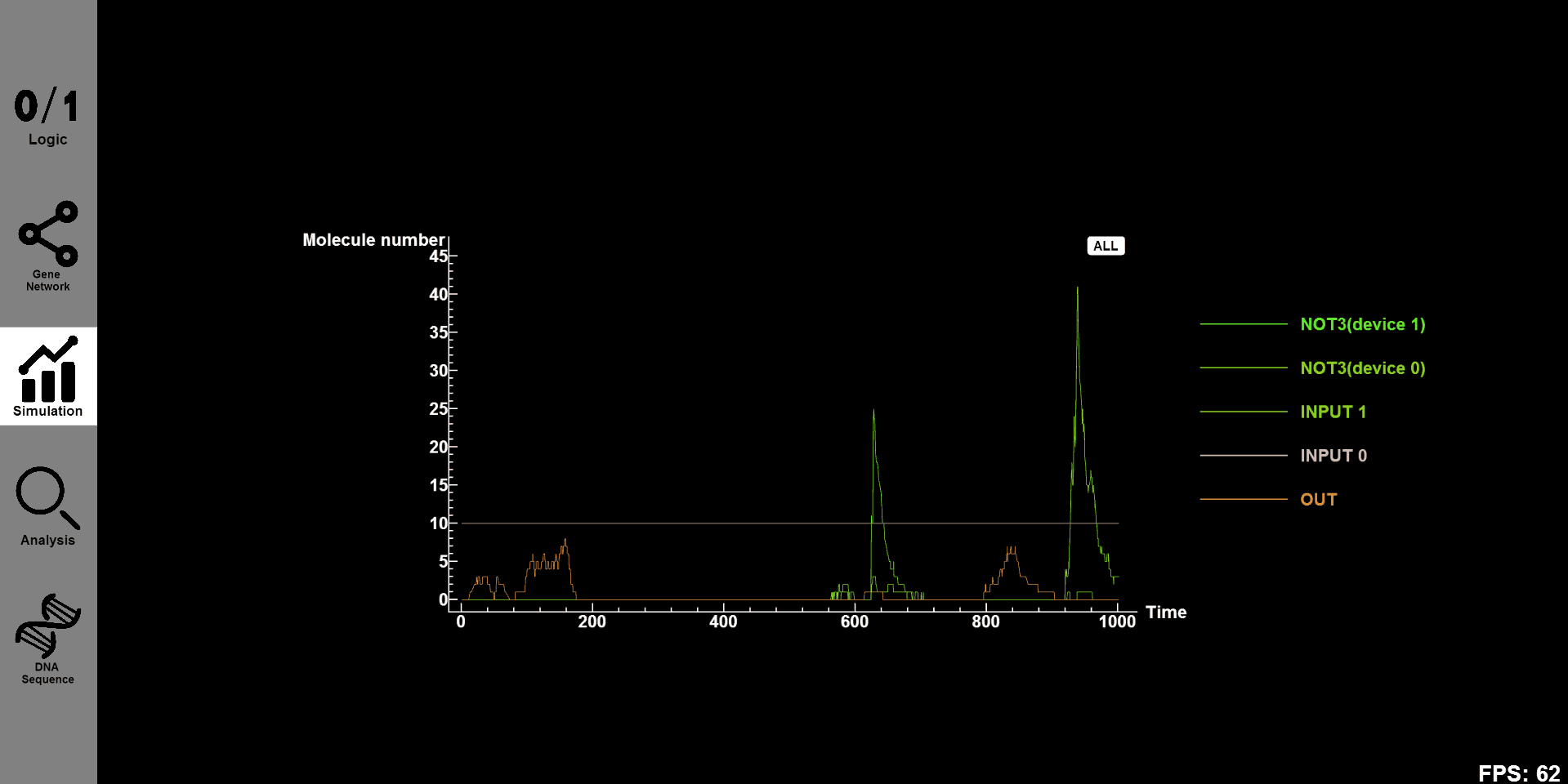
Details:
There are two formalisms for mathematically describing the time behavior of a spatially homogeneous chemical system.
The first one is the deterministic approach which regards the time evolution as a continuous and wholly predictable process governed by a set of coupled ordinary differential equations (the ¡°reaction-rate equations¡±).
The other one is the stochastic approach which regards the time evolution as a kind of random-walk process governed by a single differential-difference equation (the ¡°master equation¡±).
Some simple arguments in kinetic theory show that the stochastic formulation of chemical kinetics has a firmer physical basis than the deterministic formulation. Unfortunately, the stochastic master equation is mathematically intractable in the most cases.
There is, however, a way to make exact numerical calculations within the framework of the stochastic formulation without having to deal with the master equation directly. It is a relatively simple digital computer algorithm, which uses a rigorously derived Monte Carlo procedure to numerically simulate the time evolution of a given chemical system. Like the master equation, this ¡°stochastic simulation algorithm¡± can correctly explain the inherent fluctuations and correlations that are unfortunately ignored in the deterministic formulation.
4.Analysis: Get robustness and sensitivity analysis
Function:
While offering various functions, BioBLESS also serves as an analysis tool for designed gene circuits. Having designed a gene circuit, you can click the ¡°Analysis¡± button to reset parameters of initial concentrations and reaction rates. Meanwhile, BioBLESS will set simulation slots and carry the simulation task automatically. According to the perturbation ratio you input, such as 20% or 30%, the concentration-time curves of inputs and outputs will be shown together to shed light on the robustness of a designed gene circuit. Moreover, data processing and demonstration will help users to better understand the robustness of designed gene circuits.
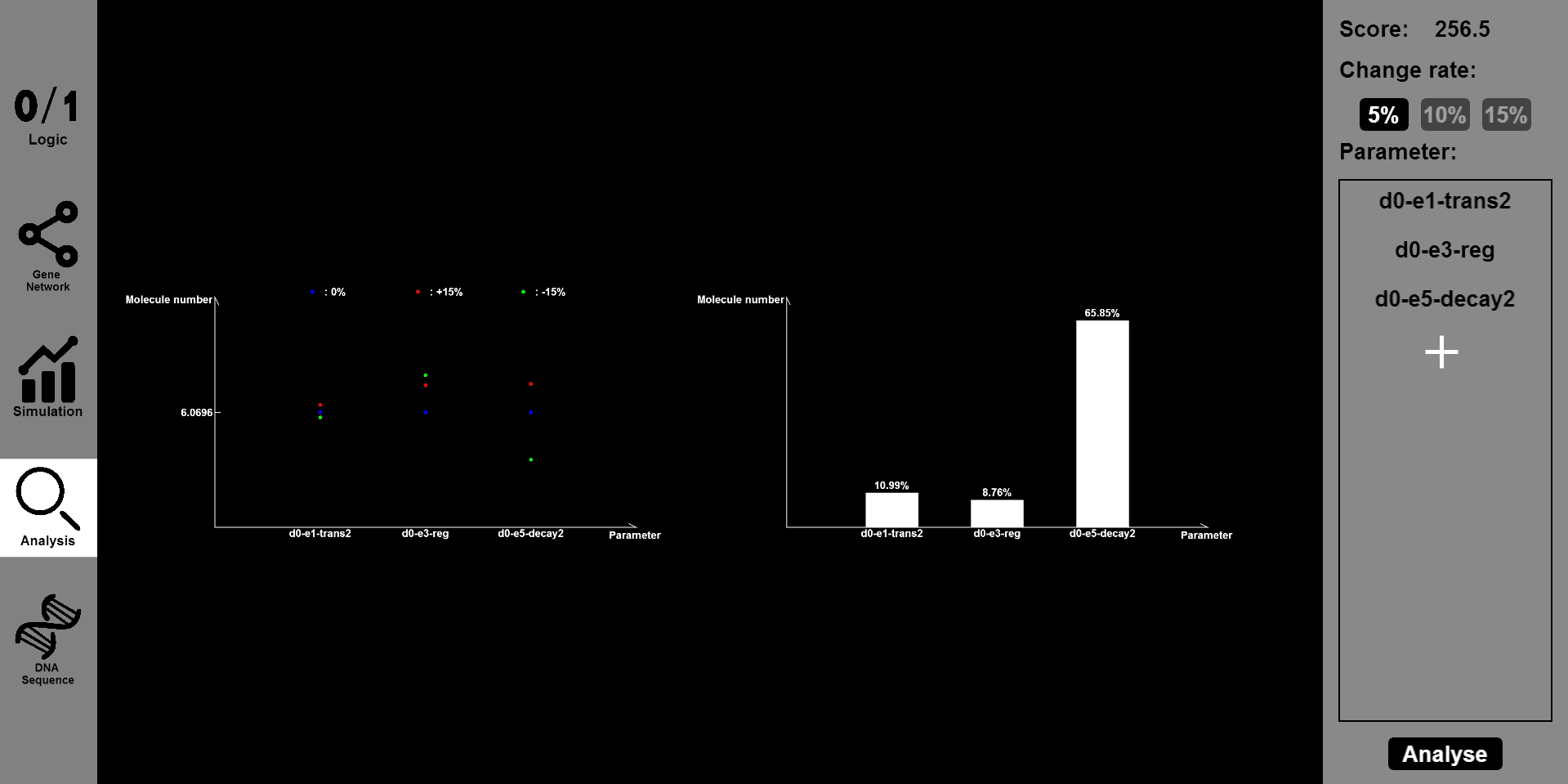
Details:
Robustness and sensitivity can measure the property of being strong and healthy or not in constitution. They refer to the ability of tolerating perturbations that might affect the system¡¯s functional body. When perturbations to the inputs of a system are small in magnitude, we hope to measure whether the concentration of outputs will change sharply or in extreme cases, even destroy the input-output logic embodied in truth tables. The most intractable problem we confront is the overload on our server to complete the simulation task. After literature reviewing, we choose some factors, for example, translation rate, to which a designed gene circuit is most sensitive. We also set a range for parameter fluctuations. Some designed circuits are proven to be robust and resistant to environment perturbations, while some other plausible well-functioning gene circuits are sensitive to specific parameters. The analysis is crucial because it gives us deeper understanding of a complicated gene circuit and lays foundation for wet-lab realization of functional novel gene circuits.
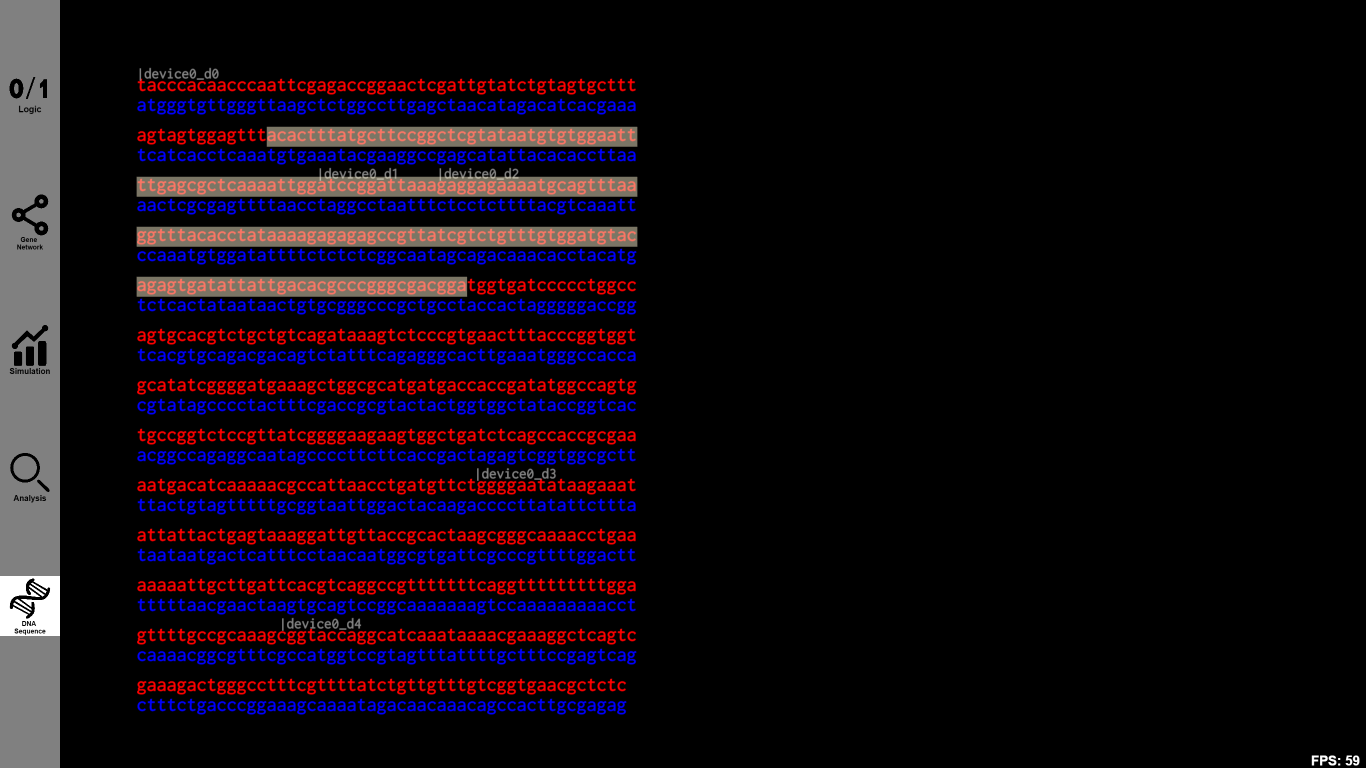
5.DNA: Get the DNA sequence
Function:
After designing gene circuits, the DNA sequence and other information of the designed circuit are available for users to check and download.
Why to choose BioBLESS
Although many designs of logic gates have come into being, circuit design in a larger scale is still rare. With more inputs and more given functions, the inter-reactions among them may become too complicated for humans to analyze one by one.
BioBLESS is here to tackle the problem.
- BioBLESS can offer multiple functions, and it can be of great help even if you need only single one function we provide. BioBLESS can generate optimized logic circuits and detailed gene circuits automatically, then depict most reliable ones for users after screening.
- BioBLESS is far more thoughtful in analyzing and displaying interior structure as well as exterior integration, which makes the simulation more accurate and reliable.
Reference
[1]Marchisio M A, Stelling J. Computational design of synthetic gene circuits with composable parts[J]. Bioinformatics, 2008, 24(17): 1903-1910.
[2]Agresti J J, Antipov E, Abate A R, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution[J]. Proceedings of the National Academy of Sciences, 2010, 107(9): 4004-4009.
[3]Alterovitz G, Ramoni M F. Systems bioinformatics: an engineering case-based approach[M]. Artech House, Inc., 2007.
[4]Batt G, Yordanov B, Weiss R, et al. Robustness analysis and tuning of synthetic gene networks[J]. Bioinformatics, 2007, 23(18): 2415-2422.
[5]Benenson Y. Biomolecular computing systems: principles, progress and potential[J]. Nature Reviews Genetics, 2012, 13(7): 455-468.
[6]Bray D, Lay S. Computer simulated evolution of a network of cell-signaling molecules[J]. Biophysical Journal, 1994, 66(4): 972-977.
[7]Burrage K, Burrage P M, Leier A, et al. Design and analysis of biomolecular circuits: engineering approaches to systems and synthetic biology[J]. 2011.
[8]Callura J M, Cantor C R, Collins J J. Genetic switchboard for synthetic biology applications[J]. Proceedings of the National Academy of Sciences, 2012, 109(15): 5850-5855.
[9]Cameron D E, Bashor C J, Collins J J. A brief history of synthetic biology[J]. Nature Reviews Microbiology, 2014, 12(5): 381-390.
[10]Eisenstein M. Microarrays: quality control[J]. Nature, 2006, 442(7106): 1067-1070.
[11]Endy D. Foundations for engineering biology[J]. Nature, 2005, 438(7067): 449-453.
[12]Ferber D. Microbes made to order[J]. Science, 2004, 303(5655): 158.
[13]Fran?ois P, Hakim V. Design of genetic networks with specified functions by evolution in silico[J]. [13]Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(2): 580-585.
[14]Ginkel M, Kremling A, Nutsch T, et al. Modular modeling of cellular systems with ProMoT/Diva[J]. Bioinformatics, 2003, 19(9): 1169-1176.
[15]Hormozdiari F, Kichaev G, Yang W Y, et al. Identification of causal genes for complex traits[J]. Bioinformatics, 2015, 31(12): i206-i213.
[16]Lv Y, Wang S, Meng F, et al. Identifying novel associations between small molecules and miRNAs based on integrated molecular networks[J]. Bioinformatics, 2015: btv417.
[17]MacDonald J T, Barnes C, Kitney R I, et al. Computational design approaches and tools for synthetic biology[J]. Integrative Biology, 2011, 3(2): 97-108.
[18]Marchisio M A, Stelling J. Automatic design of digital synthetic gene circuits[J]. PLoS Comput. Biol, 2011, 7(2): e1001083.
[19]Marchisio M A. In silico design and in vivo implementation of yeast gene Boolean gates[J]. Journal of biological engineering, 2014, 8(1): 1-15.
[20]Moon T S, Lou C, Tamsir A, et al. Genetic programs constructed from layered logic gates in single cells[J]. Nature, 2012, 491(7423): 249-253.
[21]Rinaudo K, Bleris L, Maddamsetti R, et al. A universal RNAi-based logic evaluator that operates in mammalian cells[J]. Nature biotechnology, 2007, 25(7): 795-801.
[22]Schonbrun E, Abate A R, Steinvurzel P E, et al. High-throughput fluorescence detection using an integrated zone-plate array[J]. Lab on a Chip, 2010, 10(7): 852-856.
[23]Siuti P, Yazbek J, Lu T K. Synthetic circuits integrating logic and memory in living cells[J]. Nature biotechnology, 2013, 31(5): 448-452.
[24]Weiss R, Homsy G E, Knight Jr T F. Toward in vivo digital circuits[M]//Evolution as Computation. Springer Berlin Heidelberg, 2002: 275-295.
[25]Zhang X D, Espeseth A S, Johnson E, et al. Integrating experimental and analytic approaches to improve data quality in genome-wide RNAi screens[J]. Journal of biomolecular screening, 2008.
[26]Zhou K, Doyle J C, Glover K. Robust and optimal control[M]. New Jersey: Prentice hall, 1996.
