Team:Paris Saclay/Measurement
Measurement
Contents
1st July
Rehydratation : I13504 J23117 J23106 J23101
2nd July
Transformation : I13504 J23117 J23106 J23101
3rd July
Liquide culture : I13504 J23117 J23106 J23101
8th July
First Digestion:
BBa_J23101 BBa_J23106 BBa_J23117
Mix:
10µL of our plasmid with promotor 1µL SpeI 1µL PstI 2µL buffer 10x FastDigest 6µL H2O
Second Digestion:
BBa_I13504
Mix:
10µL of our plasmid with gene 1µL XbaI 1µL PstI 2µL buffer 10x FastDigest
Incubation 1h30, 37°C
9th July
Transformation:
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
On LB + Chloramphenicol 20ug/mL. Incubation ON, 37°C
15th July
Liquid culture:
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
We can observe from the plate that BBa_J23101 + BBa_I13504 and BBa_J23106 + BBa_I13504 are yellow when we expose them to the UV light. But BBa_J23117 + BBa_I13504 don't seem to be yellow on the UV light.
16th July
Digestion:
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
Reaction mix:
Plasmid: 2µL EcoRI: 0,5µL PstI: 0,5µL Buffer FastDigest (10x): 2µL H2O: 15µL
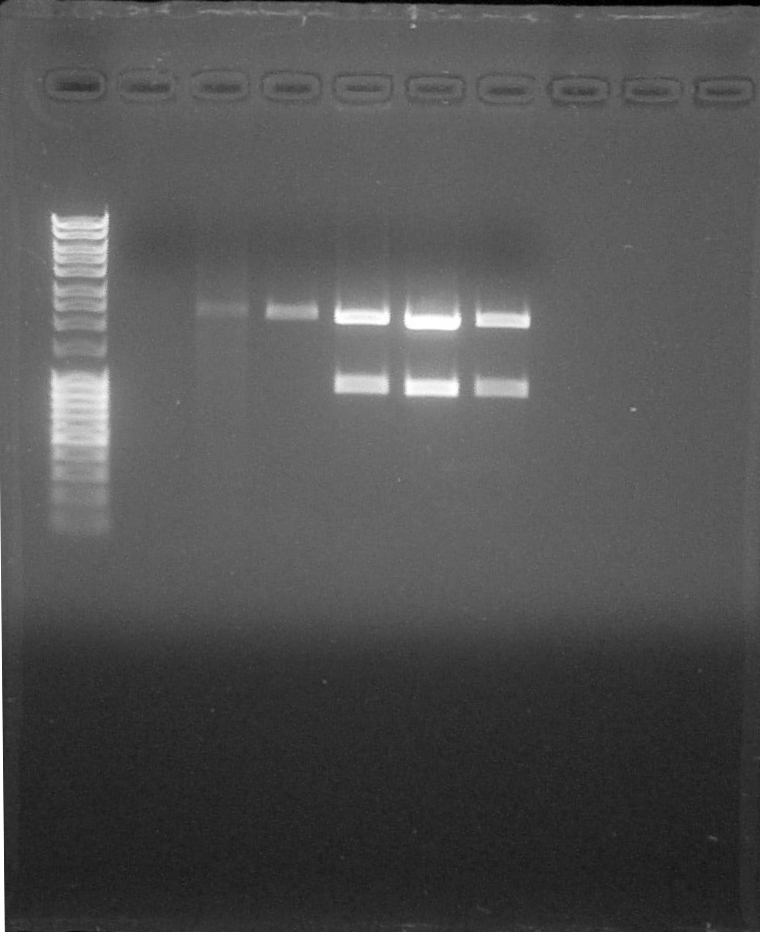
Electrophoresis:
Preparation of Agarose Gel 1%, 0,5g in 50mL of 1X TAE, 0,5µL of BET Migration 0,06A 80V
17th July
New culture on Plate
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
23rd July
Liquid culture from the 3 stocks
24th July
Cytometer
We count 500 000 events Controls:
Cells alones Cells transformed by BBa_J23101, BBa_J23106 and BBa_J23117
Our measurements: Cells transformed by
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
We uses cells in growth phase and stationary phase
Between each test, we do 2 washes with bleach and 2 washes with H2O
28th July
New culture of BBa_J23101/BBa_J23106/BBa_J23117 + BBa_I13504
Tecan utilisation:
we use only LB without chloramphenicol and we suspect a contamination of our samples. We depose in inch well 300µL We analyse the OD and fluorescence's variation (the excitation and emission wave lenght were choose after a scan in the process to obtain the best results) For each sample, we depose twelve time (12x8 plate)
- LB
- Competent cells
- Cells with J23101
- Cells with J23101 + GFP
- Cells with J23106
- Cells with J23106 + GFP
- Cells with J23117
- Cells with J23117 + GFP
We let's run for 20 cycles of 1 hour
.=29th July=
Tecan utilisation:
This time, we use LB and chloramphenicol because the plate has just a protection and we suspect a contamination of our samples. We depose in inch well 300µL We analyse the OD and fluorescence's variation (the excitation and emission wave lenght were choose after a scan in the process to obtain the best results)
For each sample we use 3 different colonies, we depose 2 times each colonies (12x8 plate)
LB Competent cells Cells with J23101 Cells with J23101 + GFP Cells with J23106 Cells with J23106 + GFP Cells with J23117 Cells with J23117 + GFP
We let's run for 20 cycles of 1 hour
Flow cytometer
We analyse the sample see previously with ODmax and OD 0.4 But we doesn't use the good scale so we will reused it tomorrow
30th July
Flow cytometer
We count 500 000 events Controls:
LB Cells transformed by BBa_J23101, BBa_J23106 and BBa_J23117
Our measurements: Cells transformed by
BBa_J23101 + BBa_I13504 BBa_J23106 + BBa_I13504 BBa_J23117 + BBa_I13504
We uses cells in growth phase and stationary phase
Between each test, we do 2 washes with bleach and 2 washes with H2O
We use a less powerful adjustment to see tall the result than the day before.