Team:UCLA/Notebook/Spider Silk Genetics/27 August 2015
Contents
8/27/2015
Sequencing Results
- M1-12(1C3)
- 1: wrong sequence entirely
- 2: wrong sequence entirely
- 3: wrong sequence entirely
- Interestingly, all three sequences align to each other.
- M1/2[1:1]-12(1C3)
- 1: Bad, single base pair mutation at ~550
- 2: GOOD
- 3: GOOD
- M1/2[1:1]-12(T7)
- 1: Bad, deletion at ~80
- 2: GOOD
- 3: possibly correct, but not trustworthy due to short read.
- M1/2[2:1]-12(1C3)
- 1: Bad, single bp mutation near ~660
- 2: Bad, single bp mutation at 962
- 3: GOOD
- M1/2[2:1]-12(T7)
- 1: bad, non-silent single bp mutation at 1004
- 2: GOOD? silent single bp mutation at 702
- 3: GOOD? silent single bp mutation at 702
Miniprep for M1/2[1:2]-12(1C3)
- Used qiagen kit to purify the two cultures.
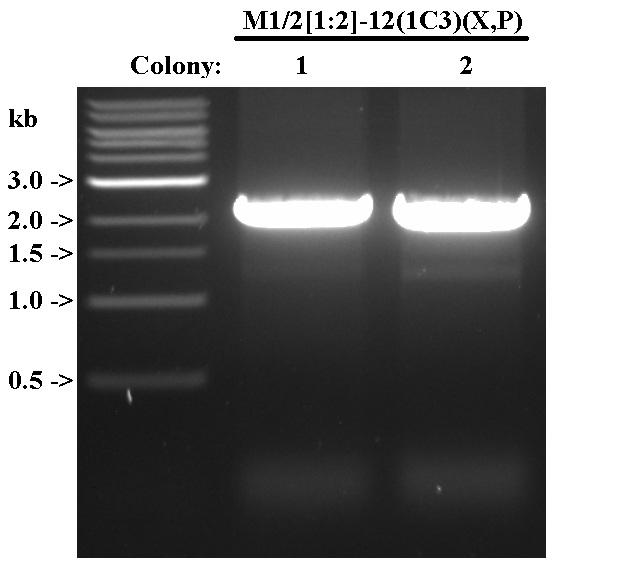
Digestion Check for M1/2[1:2]-12(1C3)
- Digest with X, P in preparation for possible subcloning.
- Digest 3 ug of each DNA in a 50 uL reaction with 1 uL each of XbaI, PstI.
- Digest at 37 C for 1.5 hrs, heat kill at 65 C for 20 min.
- Visualize on 1% gel with 2 uL of NEB 1 kb ladder.
- Both colonies were wrong; did not send for sequencing.
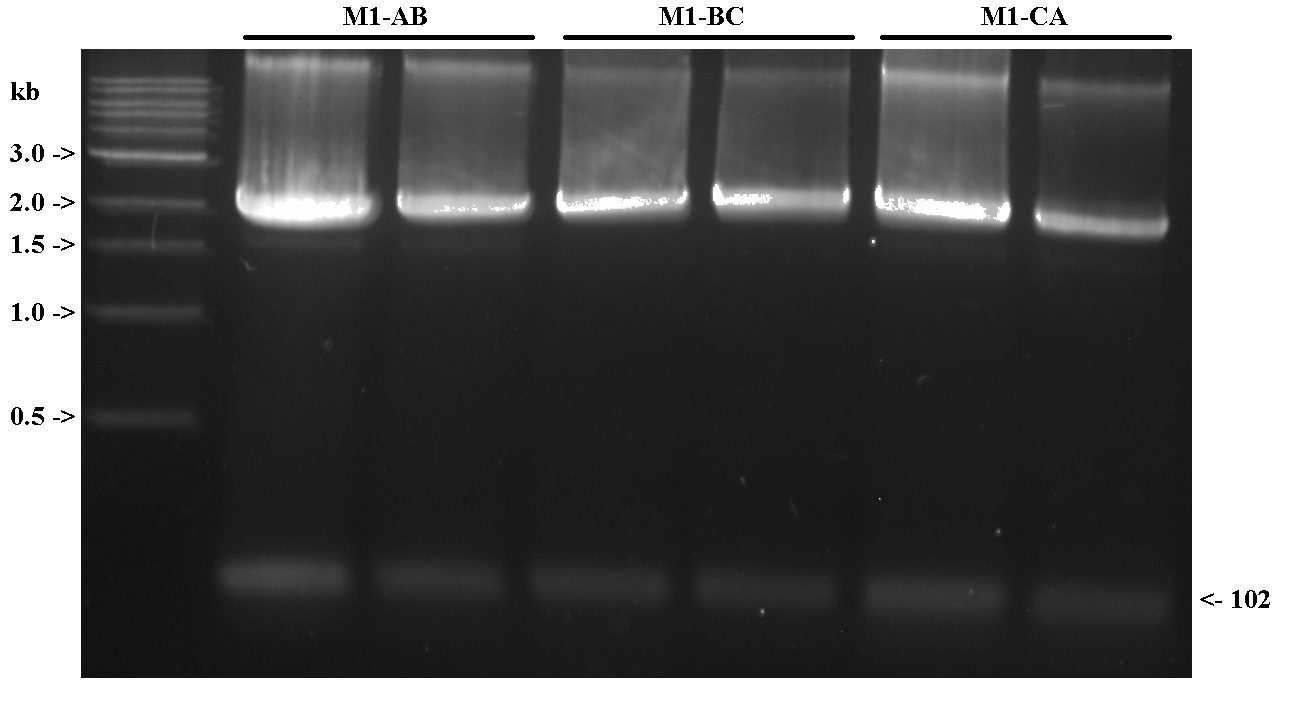
BsaI Digestion of M1 Monomers
- Digested 5 ug of plasmid DNA in a 50 uL reaction with 4 uL of BsaI. Digest 2x 50 uL reactions for each monomer.
- Digest at 50 C for 2 hrs, heat at 65 C for 20 min.
- Run of 1.5% TAE gel to visualize. Used 2 uL of NEB 1 kb ladder. (Should have used the 50 bp or 100 bp ladder instead)
- Olivia excised the gel for extraction.