Team:UCLA/Notebook/Spider Silk Genetics/6 July 2015
7/6/2015
MaSp2 Sequencing Core Assembly
- PCA for MaSp2 Sequencing Core to be used for sequencing longer MaSp constructs.
- Used the MaSp2 Seq Core 1-F (D-05) and MaSp Seq Core 1-R (E-05) to create full length MaSp sequencing core.
| Volume (uL) | |
|---|---|
| 5x Q5 Buffer | 5 |
| 10 mM dNTPs | 0.5 |
| 10 uM For (D-05) | 1.25 |
| 10 uM Rev (E-05) | 1.25 |
| 5x GC Enhancer | 5 |
| ddH2O | 11.75 |
| Q5 Polymerase | 0.25 |
| Total | 25 |
- NEB Tm calculator gives 65 C for annealing, based on the overlap between the forward and reverse.
- Test 63 C and 66 C.
| 98 C | 30 sec |
| 98 C | 10 sec |
| 63, 66 C | 15 sec |
| 72 C | 10 sec |
| repeat from step 2 | 25 x |
| 72 C | 2 min |
| 12 C | hold |
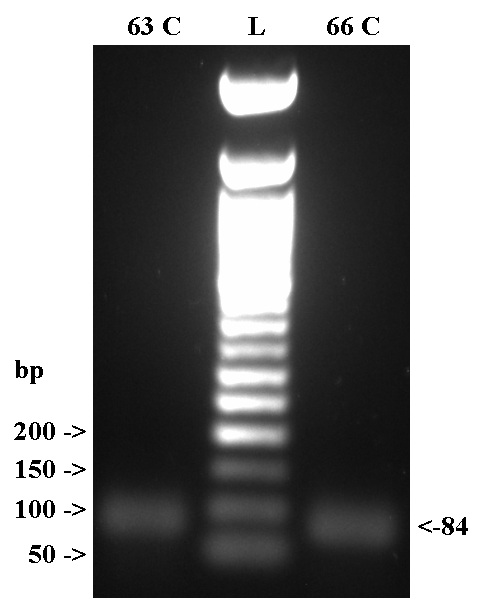
Results
- Cast 1.5 % TAE gel, used 2 uL of 50 bp ladder to visualize.
- The assembly was successful.
ICA using M-270 Beads
- The Streptavidin M-270 Dynabeads came in today. We will test ligation of a 3-mer using these beads to see if we get a different result compared to using Magnabind Streptavidin beads.
- Ligations were performed in 10 uL reactions using T7 ligase (Enzymatics), 50 ng of the relevant DNA, and 1 uL of the relevant 5 uM capping oligo.
- 5 uL of beads were used.
- Washed twice using 2x BW buffer.
- Resuspend washed beads in 5 uL 2x BW buffer, 1 uL of 5 uM initiator, and 4 uL ddH2O.
- Rotate at RT for 45 minutes.
- Wash twice using 0.5x BW buffer.
- Add ligation mix
- 5 uL 2x T7 ligase buffer
- 50 ng 2AB
- 0.5 uL TL ligase
- ddH2O to 10 uL
- Repeat ligations and washes as needed.
- Final wash using ddH2O.
- Elute in 15 uL 0.01% Tween-20 at 95 C for 3 min.
- Saved Elution.