Team:Uniandes Colombia/DryLab
Mathematical Models
We use a mathematical model to check if the design works before starting to work at the laboratory, or improve the response of the original construct. To develop this model, the first step was to create a deterministic model, which was based on differential equations. This type of model describes the mean behavior for each of the substances in the synthetic circuit over time. However, it does not take into account the probabilities involved in each of the events described, the population interactions or the noise of the system.
The deterministic model is based on the law of mass conservation and expresses the inputs and outputs of the system with expressions that use the law of mass action and known models like Hill's or Michaelis Menten.However, we only have a model that shows a response that does not take into account the stochasticity of Nature. That's why did a second simulation: The stochastic Model. You are very welcome to check it all.
Differential equations
Synechococcus elongatus
Our clock protein binds to our promoter(pCL) and then transcribes LuxI and LacI proteins. We have to take into account the degradation(g) and the diffusion coefficient(c) if it is needed. After that, the two proteins go through independent paths.
Lux I Equation
LuxI has a basal production (alpha), a degradation rate (gamma), and is positively regulated by the clock protein. This positive regulation is represented as a hill equation for activated genes, represented through the second term.
$$ \frac{dXI}{dt}=a_{s}+\frac{b_{s}*CL^{n_{s}}}{CL^{n_{s}+k_{s}^{n_{s}}}}-g_{XI}*XI$$The product of luxI catalyzes the production of AHL
Lac I Equation
The terms that describe la I are analogous to those of luxI. There is a basal production rate (alpha), a degradation rate (gamma), and a term describing lacI production due to positive regulation by the clock protein (hill equation for genes that are positively regulated).
$$ \frac{dLI}{dt}=a_{s}+\frac{b_{s}*CL^{n_{s}}}{CL^{n_{s}+k_{s}^{n_{s}}}}-g_{LI}*LI$$Intracellular AHL-degrading enzyme equation
This enzyme, encoded by the aaiA gene, has a basal production (alpha), a degradation rate (gamma), a term accounting for how the negative feedback due to lacI (hill equation for genes with a negative regulator) influences the enzyme production, and a negative term (c) due to difussion outside the cell.
$$ \frac{dEA_{i}}{dt}=a_{L}+\frac{b_{s}}{1+(\frac{LI}{k_{L}})^{n_{L}}}-g_{LI}*LI$$
Shewanella
LuxR
LuxR has a basal production (alpha), a degradation rate (gamma), a negative term due to its coupling to AHL, and a positive term due to its decoupling from AHL.
$$\frac{dXR}{dt}=a-b_{XR}*XR*A-g_{XR}*XR+d_{XRa}*XRa$$
LuxR-AHL complex
The formation of the complex depends on a formation rate (ba) and the concentrations of LuxR (XR) and AHL (A). It also has a degradation rate (gamma), and a negative term due to decoupling of the complex (at a rate dXR).
$$\frac{dXRa}{dt}=b_{XR}*XR*A-g_{XRa}*XRa+d_{XR}*XRa$$
Cytochrome Equation
Cytochromes have a basal production (alpha), a degradation rate (gamma), and a production due to the attachment of the LuxR-AHL complex to the promoter.
$$\frac{dCY}{dt}=a_{X}+k_{c}*XR*A-g_{CY}*XR$$
Deterministic model
Pclock varies sinusoidally with time. That's why a sinusoidal input was given for it.
Synechococcus elongatus
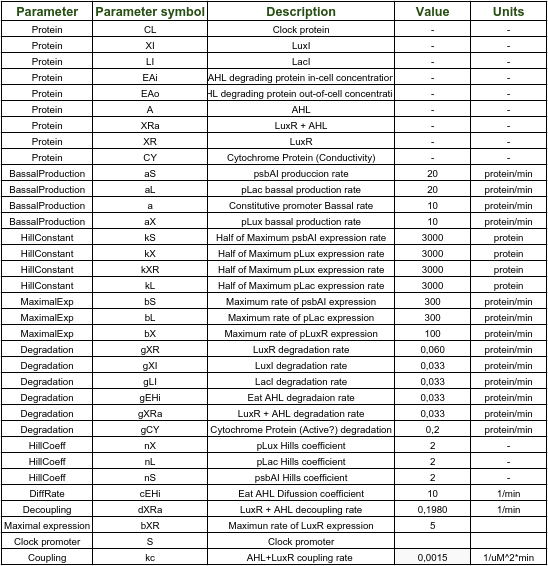
As evidenced in the graph above, AHL also behaves sinusoidally as an output. Consequently, it is logical to have a sine wave with a 24hr period as input for Shewanella oneidensis
Shewanella oneidensis
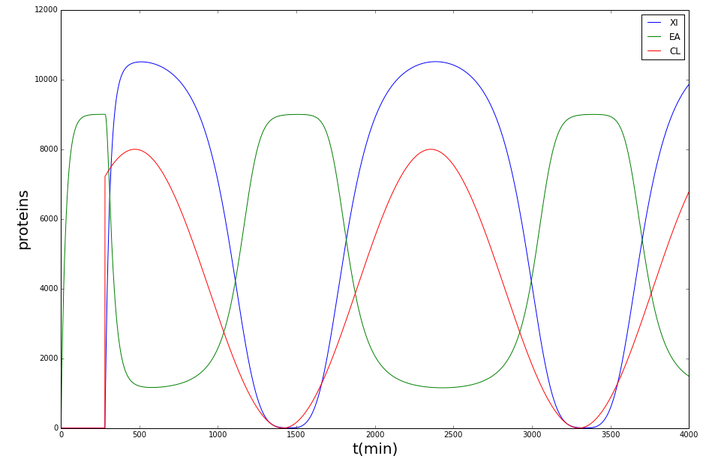
It was thus shown that with the given parameters, the cytochromes (and so the resistance) in Shewanella
Stochastic model
Until now, all we have is a deterministic model of our system. This model takes into account exact concentrations of molecules as the variables in the differential equations. This means that all the calculations were made taking into account the mean values of the concentrations of particles.
Although this is a good approach, modelling the biological processes as random, stochastic events reflects a more accurate version of reality.
The following passage by physicist Daniel Gillespie provides some insight on this claim:
“…For “ordinary” chemical systems in which fluctuations and correlations play no significant role, the method stands as an alternative to the traditional procedure of numerically solving the deterministic reaction rate equations. For nonlinear systems near chemical instabilities, where fluctuations and correlations may invalidate the deterministic equations, the method constitutes an efficient way of numerically examining the predictions of the stochastic master equation. Although fully equivalent to the spatially homogeneous master equation, the numerical…” (Gillespie 1976)
To tackle this problem, Daniel Gillespie designed the stochastic simulation algorithm. This method is particularly useful when reducing computational costs.
The algorithm is based on the reaction probability density function; a function that essentially dictates
the amount of time that will pass between events such as the creation or degradation of a protein. This time interval is given by the equation:
The algorithm is based on the reaction probability density function; a function that essentially dictates
the amount of time that will pass between events such as the creation or degradation of a protein.
To determine which event will occur, a second random number from 0 to 1 is used. After the events are normalized, depending where in the range of the event normalization the random number falls, a particular event is chosen.
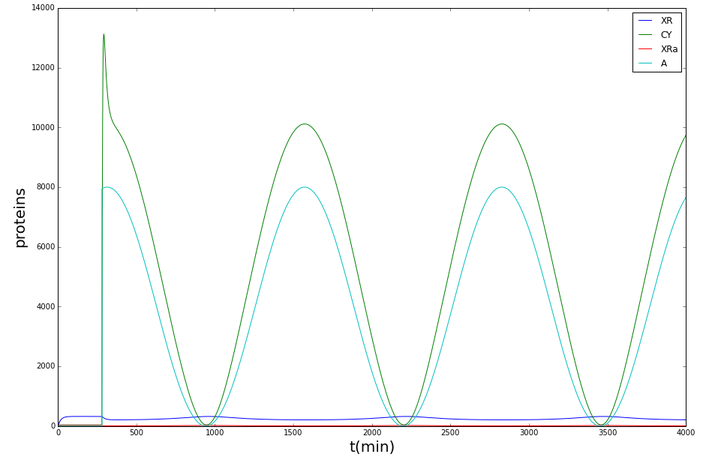
The following graphs display the outcome of the simulations for Synechococcus elongatus and Shewanella oneidensis.
Synechococcus elongatus
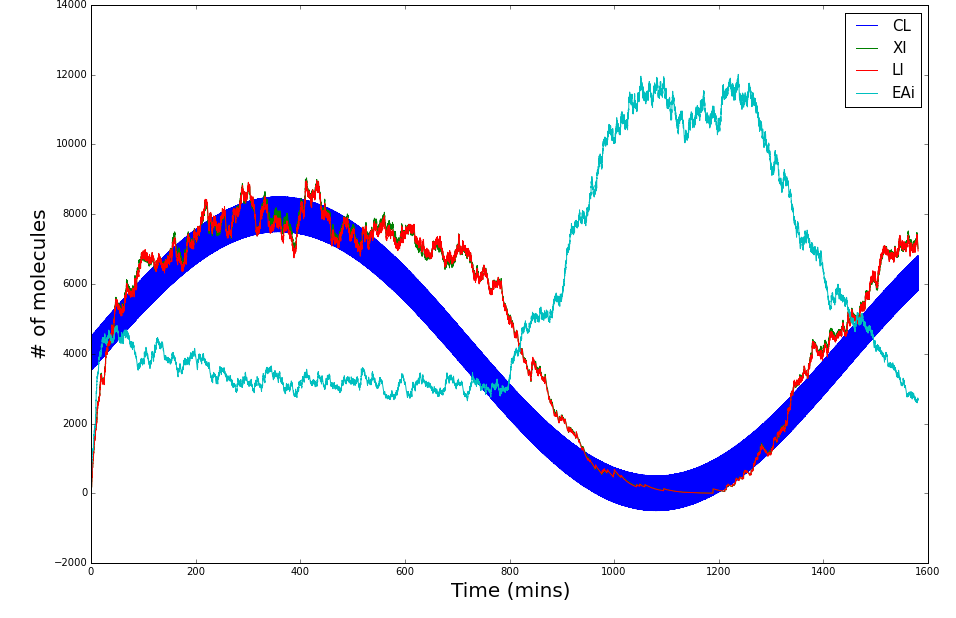
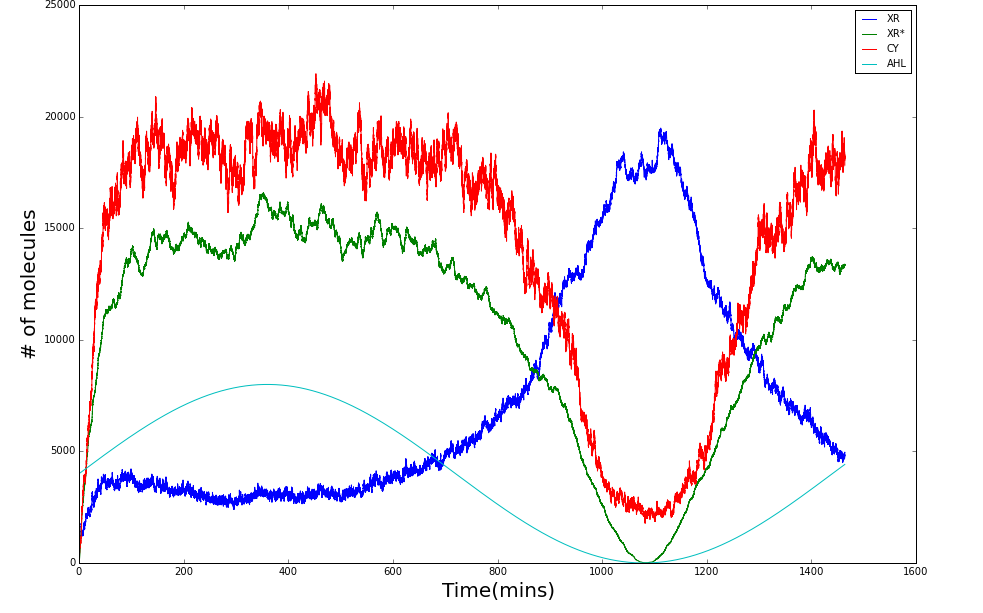
Shewanella oneidensis
Scripting
Deterministic Model
Synechococcus elongatus
Shewanella onediensis
Stochastic Model
Synechococcus elongatus
Shewanella oneidensis
Uploading pictures and files
You can upload your pictures and files to the iGEM 2015 server. Remember to keep all your pictures and files within your team's namespace or at least include your team's name in the file name.
When you upload, set the "Destination Filename" to Team:YourOfficialTeamName/NameOfFile.jpg. (If you don't do this, someone else might upload a different file with the same "Destination Filename", and your file would be erased!)