Difference between revisions of "Team:Czech Republic/Project/Synthetic haploids"
(→Key Achievements) |
CecylHynek (Talk | contribs) (→Abstract) |
||
| Line 5: | Line 5: | ||
</html> | </html> | ||
== Abstract == | == Abstract == | ||
| − | + | To use the mating pheromones as signals for multicellular communication synthetic haploids of both mating types that preserve the ability to process an extracellular signal via pheromone response pathway even after mating - in diploid state are needed. However, all components of the pathway are switched-off naturally in diploid cells, therefore synthetic haploid strains that mate into a diploid with a functional pheromone pathway were designed. As a result, the IODs can use the robust natural signaling pathway for signal transduction. | |
<html> | <html> | ||
</div> | </div> | ||
Revision as of 19:58, 14 September 2015
Signal transduction
Introduction
Abstract
To use the mating pheromones as signals for multicellular communication synthetic haploids of both mating types that preserve the ability to process an extracellular signal via pheromone response pathway even after mating - in diploid state are needed. However, all components of the pathway are switched-off naturally in diploid cells, therefore synthetic haploid strains that mate into a diploid with a functional pheromone pathway were designed. As a result, the IODs can use the robust natural signaling pathway for signal transduction.
Key Achievements
- Constructed a set of reporter promoters for yeast cells.
- Characterized reporter promoters
- Created synthetic MATa and MATx strains
- Built a synthetic diploid strain with a functional yeast pheromone pathway
- Demonstrated the correct functionality of yeast pheromone pathway cascade in synthetic diploids
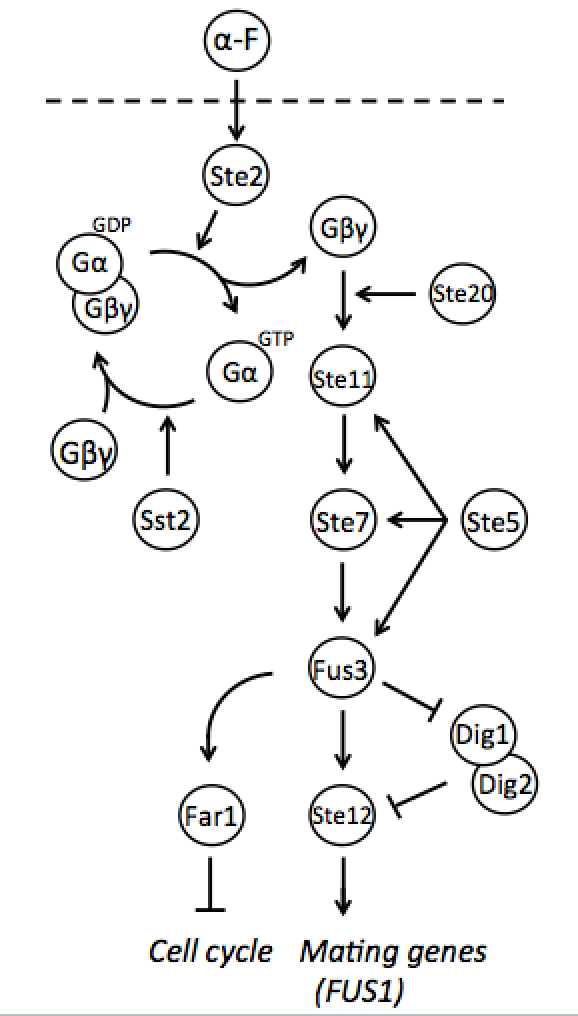
Yeast pheromone pathway
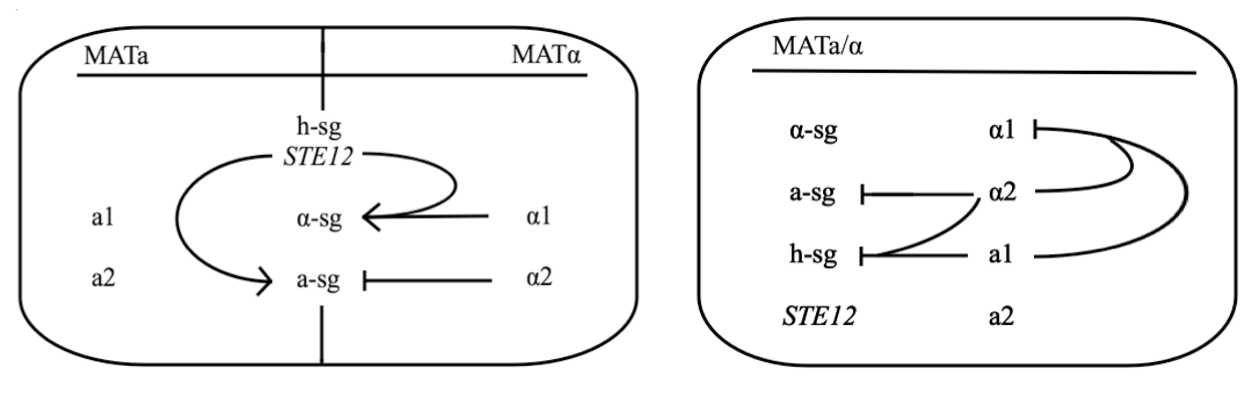
Yeast Saccharomyces cerevisiae exists either in haploid or diploid state. The two mating types are called MATa and MATα and differ only within approx. 2 kbp long region on chromosome III called MAT locus. MATa locus expresses transcription factors a1 and a2, whereas MATα locus expresses transcription factors α1 and α2. Both types express three groups of genes, which are:
- haploid-specific genes (h-sg)
- a-specific genes (a-sg)
- α-specific genes (α-sg)
As their names indicate, these genes are only active in haploids, MATa cells, or MATα cells, respectively.
Both mating types constantly produce small amounts of mating type-specific pheromone and when the two cells of opposite types are in close proximity they identify each other by sensing each other’s pheromone. The response to detected pheromone is facilitated by so called pheromone pathway, which is a cascade of chemical reactions that results in the preparation for mating. Since only haploids mate, the yeast pheromone pathway is only functional in haploid cells.
Design
Reconstruction of yeast pheromone pathway in diploid cell
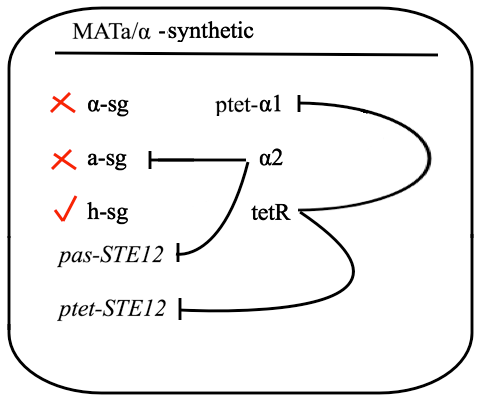
The schematic shows that h-sg genes are switched-off in diploid cell and since all genes included in yeast pheromone pathway signal transduction (except from the pheromone receptor)are h-sg, the pathway can be reconstructed by switching them on. It can be managed by interrupting one of the h-sg regulators - a1 or α2. While a1 has no function in haploid, its deletion has no effect in haploid cell, but in diploid it interrupts the repression of h-sg.
Naturally in haploids, after exposure to mating pheromone yeast pheromone pathway results in induction of expression of mating genes that initiate the mating process. Since it is not desired for the synthetic diploid to mate, the initiation of mating process needs to be disrupted. It can be achieved by repressing Ste12 transcription in the diploid. While Ste12 is fundamental in both haploid types in the process of mating, the designed mechanism must preserve transcription in haploid cells and repress it in diploid.
For this purpose, mechanism of tetracycline-controlled transcriptional activation was used: a1 gene in MATa was replaced by TetR and STE12 was put under the control of synthetic a-specific promoter that is repressed in the same way as a-sg. In MATα, α1 repression was preserved by placing it under the control of pTet and STE12 was also put under the control of pTet. As a result, transcription of Ste12 is preserved in both haploids and repressed in diploid - pTet-STE12 thanks to TetR repressor that is expressed from MATa chromosome, and a-specific STE12 thanks to α2. Also h-sgare expressed because of disruptedrepression by deleting a1 preserving transcription of all the genes forming yeast pheromone pathway.
See following pages for detailed information about constructing the synthetic haploids:
Materials and methods
Used strains
- Ecoli E5alpha
- 7283 MATx yeast strain
- 7284 MATa yeast strain
- 10150 MATa/x yeast strain
- 6193 MATa yeast strain (URA LEU HIS)
- 6194 MATx yeast strain (URA LEU TRP)
Used material
- LB-M agar plates with chloramphenicol
- LB-M agar plates with ampicillin
- 1.5 ml eppendorf tubes
- 0.5 ml PCR tubes
- 50 ml centrifuge conical base and rim tubes
- NucleoSpin Plasmid DNA, RNA, and protein purification Kit
- NucleoSpin Gel and PCR Clean-up Kit
- LB-M medium with chloramphenicol
- NaOH agarose gel and buffer
- Sphero Rainbow Calibration Particles, 8 Peaks, 3.0-3.4
Used methods
- Transformation
- Miniprep
- Restriction digest
- Ligation
- NucleoSpin Gel Clean-up
- NucleoSpin Plasmid DNA purification
- Flow cytometry
All used protocols can be found here:
Protocols
Used software
- CFlow Plus
- Microsoft Excel
- Sphero PMT QC Template
Used Parts
yG_MATa :
AATTCATCTAGAGAAGAAAGCAAAGCCTTAATTCCAAGGAAAAAGAAGAAGTTGCAAAGAAATGTGGCATTACTCCACTTCAAGTAAGAGTTTGGGTATGTAATATGAGAATCAAACTTAAATATATCCTATACGTAGTATGGCGGAAAACATAAACAGAACTCTGTTTAACATTCTAGGTACTGAGcaaattaaagccttcgagcgtcccaaaaccttctcaagcaaggttttcagtataatgttacatgcgtacacgcgtctgtacagaaaaaaaagaaaaatttgaaatataaataacgttcttaatactaacataactataaaaaaataaatagggacctagacttcaggttgtctaactccttccttttcggttagagcggatgtggggggagggcgtgaatgtaagcgtgacataactaatCTAAAATTCCCGGGATCCGCTGTACGCGGACCCACTTTCACATTTAAGTTGTTTTTCTAATCCGCATATGATCAATTCAAGGCCGAATAAGAAGGCTGGCTCTGCACCTTGGTGATCAAATAATTCGATAGCTTGTCGTAATAATGGCGGCATACTATCAGTAGTAGGTGTTTCCCTTTCTTCTTTAGCGACTTGATGCTCTTGATCTTCCAATACGCAACCTAAAGTAAAATGCCCCACAGCGCTGAGTGCATATAATGCATTCTCTAGTGAAAAACCTTGTTGGCATAAAAAGGCTAATTGATTTTCGAGAGTTTCATACTGTTTTTCTGTAGGCCGTGTACCTAAATGTACTTTTGCTCCATCGCGATGACTTAGTAAAGCACATCTAAAACTTTTAGCGTTATTGCGTAAAAAATCTTGCCAGCTTTCCCCTTCTAAAGGGCAAAAGTGAGTATGGTGCCTATCTAACATTTTAATAAGTTGATTGTATGCTTGGTATAGCTTGAAATATTGTGCAGAAAAAGAAACAAGGAAGAAAGGGAACGAGAACAATGACGAGGAAACAAAAGATTAATAATTGCAGGTCTATTTATACTTGATAGCAAGACAGCAAACTTTTTTTTATTTCAAATTCAAGTAACTGGAAGGAAGGCCGTATACCGTTGCTCATTAGAGAGTAGTGTGCGTGAATGAAGGAAGGAAAAAGTTTCGTGTGCTTCGAGATACCCCTCATCAGCTCTGGAACAACGACATCTGTTGGTGCTGTCTTTGTCGTTAATTTTTTCCTTTAGTGTCTTCCATCATTTTTTTGTCATTGCGGATATGGTGAGACAACAACGGGGGAGAGAGAAAAGAAAAAAAAAGAAAAGAAGTTGtaaacccacaccgggtgtcataatcaaccaatcgtaacttcatctcttccacccatgtctctttgagcaataaagccgataacaaaatctttgtcgctcttcgcaatgtcaacagtacccttagtatattctccagtagatagggagcccttgcatgacaattctgctaacatcaaaaggcctctaggttcctttgttacttcttctgccgcctgcttcaaaccgctaacaatacctggTccACTAGTCCCGGGAGCAAGATCAAGATGTTTTCACCGATCTTTCCGGTCTCTTTGGCCGGGGTTTACGGACGATGGCAGAAGACCAAAGCGCCAGTTCATTTGGCGAGCGTTGGTTGGTGGATCAAGCCCACGCGTAGGCAATCCTCGCAGATCTCGAACCATGTAATTTCCGAATACGGTAATTACACGCATCGAGCAGATCCGCCAGGCGTGTATATATAGCGTGGATGGCCAGGCAACTTTAGTGCTGACACATACAGGCATATATATATGTGTGCGACGACACATGATCATATGGCATGCATGTGCTCTGTATGTATATAAAACTCTTGTTTTCTTCTTTTCTCTAAATATTCTTTCCTTATACATTAGGACCTTTGCAGCATAAATTACTATACTTCTATAGACACACAAACACAAATACACACACTAAAaagctt
yG_MATx1 :
AAGCTTGGATTCTCACAATCCTGTCGGTCACTTCTCGGCTGTTCGCGTATATTTTTTGTTGATACTTTTACCGGTATTTTGTCTGTAATTTATTCTCTATCACTGATAGGGACTTCTCTATCACTGATAGGGAACCCAGCCTGATTTATACTATTAGGGATCGCAGGAAGGCGGTGGGAAGTCCGGGAGTCGCTGAGGGGAAGTGTCAGTGGTTTTGGGTATAAATGGCTGGTTGTTCCCTATCAGTAATAGAGAATTCCCTATCAGTGATAGAGACTGCGGATTTAGAAACTACCTGATAAAAGTATCAACAAAAATTGCGCATGCCGGCCTGGATTTTGCGCAAATTTACCTTAACGTCCCACAATATGTTTACTTCGAAGCCTGCTTTCAAAATTAAGAACAAAGCATCCAAATCATACAGAAACACAGCGGTTTCAAAAAAGCTGAAAGAAAAACGTCTAGCTGAGCATGTGAGGCCAAGCTGCTTCAATATTATTCGACCACTCAAGAAAGATATCCAGATTCCTGTTCCTTCCTCTCGATTTTTAAATAAAATCCAAATTCACAGGATAGCGTCTGGAAGTCAAAATACTCAGTTTCGACAGTTCAATAAGACATCTATAAAATCTTCAAAGAAATATTTAAACTCATTTATGGCTTTTAGAGCATATTACTCACAGTTTGGCTCCGGTGTAAAACAAAATGTCTTGTCTTCTCTGCTCGCTGAAGAATGGCACGCGGACAAAATGCAGCACGGAATATGGGACTACTTCGCGCAACAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAGTGGTTGACGAATAATTATGCTGAAGTACGTGGTGACGGATATTGGGAAGATGTGTTTGTACATTTGGCCTTATAGAGTGTGGTCGTGGCGGAGGTTGTTTATCTTTCGAGTACTGAATGTTGTCAGTATAGCTATCCTATTTGAAACTCCCCATCGTCTTGCTGCAG
yG_MATx2 :
CTGCAGAGTAGTGTCTGAGGTACAAACATCTTAGTAGTGTCGAGAGGGTTGATTGTTTATGTATTTTTGCGAAATATATATATATATATTCTACACAGATATATACATATTTGTTTTTCGGGCTCATTCTTTCTTCTTTGCCAGAGGCTCACCGCTCAAGAGGTCCGCTAATTCTGGAGCGATTGTTATTGTTTTTTCTTTTCTTCTTCTATTCGAAACCCAGTTTTTGATTTGAATGCGAGATAAACTGGTATTCTTCATTAGATTCTCTAGGCCCTTGGTATCTAGATATGGGTTCTCGATGTTCTTTGCAAACCAACTTTCTAGTATTCGGACATTTTCTTTTGTAAACCGGTGTCCTCTGTAAGGTTTAGTACTTTTGTTTATCATATCTTGAGTTACCACATTAAATACCAACCCATCCGCCGATTTATTTTTCTGTGTAAGTTGATAATTACTTCTATCGTTTTCTATGCTGCGCATTTCTTTGAGTAATACAGTAATGGTAGTAGTGAGTTGAGATGTTGTTTGCAACAACTTCTTCTCCTCATCACTAATCTTACGGTTTTTGTTGGCCCTAGATAAGAATCCTAATATATCCCTTAATTCAACTTCTTCTTCTGTTGTTACACTCTCTGGTAACTTAGGTAAATTACAGCAAATAGAAAAGAGCTTTTTATTTATGTCTAGTATGCTGGATTTAAACTCATCTGTGATTTGTGGATTTAAAAGGTCTTTAATGGGTATTTTATTCATTTTTTCTTAGTGTGTGTATTTGTATTTGCGTGTCTATAGAAGTATAGTAATTTATGCTGCAAAGGTCCTAATGTATAAGGAAAAAAAATTTAGAGAAAAAAAGAAAAAAAGAGTTTTATATACATACAGAGCACATACATGCCATATAATCATGTATATACGCGCACATATATATATGCCTGTATGTGTCAGCACTAAATTTACCTGAACATACGCGCTATATATACGCGCCTCGCGTATATGCTCGAGGATTCCCTACGCGTGGGCTTTTTTTACTAACCAACGCGCGCGAAATActagt
Construction
Construction of reporter plasmids
The pADH1, pSTE2, pSTE5, and pFUS1 promoters were obtained by PCR from yeast genome (isolated according to standard protocol from 7283 MATx strain). The asCYC1 and pTv3 promoters were obtained by PCR from g-blocks. The primers used for this are as follows:
INSERT TABLE OF PRIMERS
All promoters were amplified in a single PCR run, with the following conditions:
| Property | Value |
|---|---|
| Polymerase | Q5 |
| Extension Time | 90s |
PCR products were then gel verified
INSERT PHOTO_1
and were then purified (insert purification kit name) and restricted by corresponding restriction enzymes, which are listed in the following table
INSERT TABLE OF RESTRICTION ENZYMES
The corresponding vector for reporter plasmids was prepared by restriction and ligation of yeGFP and CYC1 terminator into a pRS416 CEN plasmid.
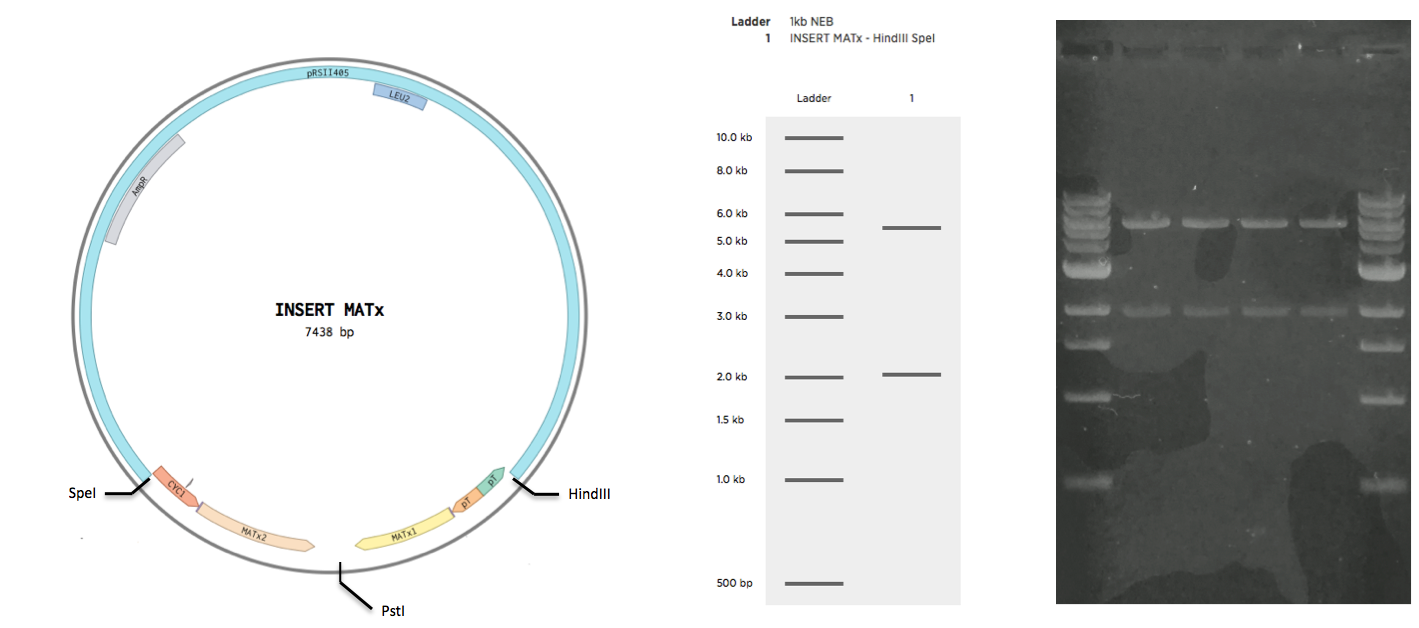
Construction of INSERT MATa and INSERT MATx
The sequence of INSERT MATa was ordered as a single gblock yG_MATa with restriction sites at the ends. The sequence of INSERT MATx was ordered as two gblocks, yG_MATx_1 and yG_MATx_2. Upon arrival, the g-blocks were resuspended in TE buffer according to the protocol of the supplier (IDT). All g-blocks were to be cloned to the backbone pRSII415 (obtained from Addgene). To achieve this the g-blocks were restricted by the following enzymes, along with the vector
| Part | 5' Enzyme | 3' Enzyme |
|---|---|---|
| yG_MATa | EcorI | HindIII |
| yG_MATx_1 | HindIII | PstI |
| yG_MATx_2 | PstI | SpeI |
| pRSII415(MATa) | EcorI | HindIII |
| pRSII415(MATx) | SpeI | HindIII |
The restrictions were all 50 ul reactions with 500 ng of DNA run with standard protocol. Enzymes and the restriction buffer were obtained from NEB. Restrictions were purified (insert purification kit name) and ligated together overnight for 16 hours at 16C. Ligations were then inactivated at 65C for 15 mins. Inactivated ligations were transformed to EC E5alpha cells. After 30 min incubation at 37C, 400 ul of each transformation was plated on a separate LB-M plate supplied with Ampicillin resistance. Plates were left overnight at 37C. The following day, patches were streaked from four from the INSERT MATa plate, and from four colonies from the INSERT MATx plate. Patches were plated on separate LB-M Ampicillin plates for INSERT MATa and INSERt MATx respectively. Plates with patches were left overnight at 37C. The following day, plasmid DNA was obtained from the patches using a miniprep kit(). Obtained plasmids were gel verified (see Validation).
Results
Final constructs
Herein we should describe the final constructs we have obtained, and add verification gels etc.
Validation of reporter plasmids
Validation of INSERT MATa and INSERT MATx
Plasmid DNA obtained from INSERT MATa-pRSII415 was restricted by EcorI and HindIII enzymes (NEB) in a CutSmart Buffer(NEB) for 60 min at 37C. Plasmid DNA obtained from INSERT MATx-pRSII415 was restricted by SpeI and HindIII enzymes (NEB) in a CutSmart Buffer(NEB) for 60 min at 37C. Restrictions were loaded in an NaOH gel. The gel was run for 90 mins at 90V.
Reporter plasmids validation
Test of mating types
Appendix
Personnel
Hynek Kasl - Responsible person
Anna Sosnová - Experimental assistance
Václav Pelíšek - Experimental assistance
Tereza Puchrová - Scientific advisor
Useful Links
Protocols page:
References
- ↑ Lin, C.-H., Choi, a., & Bennett, R. J. (2011). Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species. Molecular Biology of the Cell, 22(24), 4918–4930. doi:10.1091/mbc.E11-09-0749