Difference between revisions of "Team:Bielefeld-CeBiTec/Results/HeavyMetals"
| Line 5: | Line 5: | ||
<head> | <head> | ||
<script> | <script> | ||
| − | $("# | + | $("#project").addClass("navbar-active"); |
| + | |||
| + | $(document).ready(function(){ | ||
| + | $("#showarsenic").click(function(){ | ||
| + | $("#arsenic").slideToggle("slow"); | ||
| + | $("#nickel").hide(); | ||
| + | $("#chromium").hide(); | ||
| + | $("#copper").hide(); | ||
| + | $("#lead").hide(); | ||
| + | $("#mercury").hide(); | ||
| + | $("#arsenicgreen").fadeToggle("slow"); | ||
| + | $("#nickelgreen").hide(); | ||
| + | $("#chromiumgreen").hide(); | ||
| + | $("#coppergreen").hide(); | ||
| + | $("#leadgreen").hide(); | ||
| + | $("#mercurygreen").hide(); | ||
| + | }); | ||
| + | |||
| + | $("#showchromium").click(function(){ | ||
| + | $("#chromium").slideToggle("slow"); | ||
| + | $("#nickel").hide(); | ||
| + | $("#arsenic").hide(); | ||
| + | $("#copper").hide(); | ||
| + | $("#lead").hide(); | ||
| + | $("#mercury").hide(); | ||
| + | $("#chromiumgreen").fadeToggle("slow"); | ||
| + | $("#nickelgreen").hide(); | ||
| + | $("#arsenicgreen").hide(); | ||
| + | $("#coppergreen").hide(); | ||
| + | $("#leadgreen").hide(); | ||
| + | $("#mercurygreen").hide(); | ||
| + | }); | ||
| + | |||
| + | $("#showcopper").click(function(){ | ||
| + | $("#copper").slideToggle("slow"); | ||
| + | $("#nickel").hide(); | ||
| + | $("#arsenic").hide(); | ||
| + | $("#chromium").hide(); | ||
| + | $("#lead").hide(); | ||
| + | $("#mercury").hide(); | ||
| + | $("#coppergreen").fadeToggle("slow"); | ||
| + | $("#nickelgreen").hide(); | ||
| + | $("#arsenicgreen").hide(); | ||
| + | $("#chromiumgreen").hide(); | ||
| + | $("#leadgreen").hide(); | ||
| + | $("#mercurygreen").hide(); | ||
| + | }); | ||
| + | |||
| + | $("#showmercury").click(function(){ | ||
| + | $("#mercury").slideToggle("slow"); | ||
| + | $("#arsenic").hide(); | ||
| + | $("#chromium").hide(); | ||
| + | $("#copper").hide(); | ||
| + | $("#lead").hide(); | ||
| + | $("#nickel").hide(); | ||
| + | $("#mercurygreen").fadeToggle("slow"); | ||
| + | $("#arsenicgreen").hide(); | ||
| + | $("#chromiumgreen").hide(); | ||
| + | $("#coppergreen").hide(); | ||
| + | $("#leadgreen").hide(); | ||
| + | $("#nickelgreen").hide(); | ||
| + | }); | ||
| + | |||
| + | $("#shownickel").click(function(){ | ||
| + | $("#nickel").slideToggle("slow"); | ||
| + | $("#arsenic").hide(); | ||
| + | $("#chromium").hide(); | ||
| + | $("#copper").hide(); | ||
| + | $("#lead").hide(); | ||
| + | $("#mercury").hide(); | ||
| + | $("#nickelgreen").fadeToggle("slow"); | ||
| + | $("#arsenicgreen").hide(); | ||
| + | $("#chromiumgreen").hide(); | ||
| + | $("#coppergreen").hide(); | ||
| + | $("#leadgreen").hide(); | ||
| + | $("#mercurygreen").hide(); | ||
| + | }); | ||
| + | |||
| + | $("#showlead").click(function(){ | ||
| + | $("#lead").slideToggle("slow"); | ||
| + | $("#arsenic").hide(); | ||
| + | $("#chromium").hide(); | ||
| + | $("#copper").hide(); | ||
| + | $("#nickel").hide(); | ||
| + | $("#mercury").hide(); | ||
| + | $("#leadgreen").fadeToggle("slow"); | ||
| + | $("#arsenicgreen").hide(); | ||
| + | $("#chromiumgreen").hide(); | ||
| + | $("#coppergreen").hide(); | ||
| + | $("#nickelgreen").hide(); | ||
| + | $("#mercurygreen").hide(); | ||
| + | }); | ||
| + | }); | ||
| + | |||
</script> | </script> | ||
</head> | </head> | ||
| Line 41: | Line 134: | ||
| + | <div style="position: relative"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/7/71/Bielefeld-CeBiTec_heavymetals_teststrip_circle.png" style="position: absolute; top: 56px; left: 49px; width: 70px" id="showarsenic"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/7/71/Bielefeld-CeBiTec_heavymetals_teststrip_circle.png" style="position: absolute; top: 56px; left: 139px; width: 70px" id="showchromium"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/7/71/Bielefeld-CeBiTec_heavymetals_teststrip_circle.png" style="position: absolute; top: 54px; left: 230px; width: 70px" id="showcopper"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/7/71/Bielefeld-CeBiTec_heavymetals_teststrip_circle.png" style="position: absolute; top: 52px; left: 315px; width: 70px" id="showlead"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/7/71/Bielefeld-CeBiTec_heavymetals_teststrip_circle.png" style="position: absolute; top: 45px; left: 400px; width: 70px" id="showmercury"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/7/71/Bielefeld-CeBiTec_heavymetals_teststrip_circle.png" style="position: absolute; top: 38px; left: 482px; width: 70px" id="shownickel"> | ||
| + | |||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/b/b8/Bielefeld-CeBiTec_heavymetals_teststrip_circlegreen.png" style="position: absolute; top: 56px; left: 49px; width: 70px; display: none" id="arsenicgreen"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/b/b8/Bielefeld-CeBiTec_heavymetals_teststrip_circlegreen.png" style="position: absolute; top: 56px; left: 139px; width: 70px; display: none" id="chromiumgreen"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/b/b8/Bielefeld-CeBiTec_heavymetals_teststrip_circlegreen.png" style="position: absolute; top: 54px; left: 230px; width: 70px; display: none" id="coppergreen"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/b/b8/Bielefeld-CeBiTec_heavymetals_teststrip_circlegreen.png" style="position: absolute; top: 52px; left: 315px; width: 70px; display: none" id="leadgreen"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/b/b8/Bielefeld-CeBiTec_heavymetals_teststrip_circlegreen.png" style="position: absolute; top: 45px; left: 400px; width: 70px; display: none" id="mercurygreen"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/b/b8/Bielefeld-CeBiTec_heavymetals_teststrip_circlegreen.png" style="position: absolute; top: 38px; left: 482px; width: 70px; display: none" id="nickelgreen"> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2015/0/08/Bielefeld-CeBiTec_heavymetals_teststrip.png" style="width: 600px; min-width: 600px" alt="teststrip"> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div id="arsenic" style="display: none"> | ||
<h1>Arsenic</h1> | <h1>Arsenic</h1> | ||
<p>We choose to work with the chromosomal arsenic operon of <i>E. coli</i>, which was used by the team from Edinburgh in 2006. This operon encodes an efflux pump which confers resistance against arsenic. The expression is controlled by the repressor ArsR, which negatively autoregulates its own expression. As<sup>III</sup> can bind to three cysteine residues in ArsR. The resulting conformational change deactivates the repressor (<a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Results/HeavyMetals#Chen2014">Chen, Rosen 2014</a>).</p> | <p>We choose to work with the chromosomal arsenic operon of <i>E. coli</i>, which was used by the team from Edinburgh in 2006. This operon encodes an efflux pump which confers resistance against arsenic. The expression is controlled by the repressor ArsR, which negatively autoregulates its own expression. As<sup>III</sup> can bind to three cysteine residues in ArsR. The resulting conformational change deactivates the repressor (<a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Results/HeavyMetals#Chen2014">Chen, Rosen 2014</a>).</p> | ||
| Line 69: | Line 194: | ||
<p id="Chen2014">Chen, Jian; Rosen, Barry P. (2014): Biosensors for inorganic and organic arsenicals. In Biosensors 4 (4), pp. 494–512. DOI: 10.3390/bios4040494.</p> | <p id="Chen2014">Chen, Jian; Rosen, Barry P. (2014): Biosensors for inorganic and organic arsenicals. In Biosensors 4 (4), pp. 494–512. DOI: 10.3390/bios4040494.</p> | ||
<p id="Karig2012">Karig, David K.; Iyer, Sukanya; Simpson, Michael L.; Doktycz, Mitchel J. (2012): Expression optimization and synthetic gene networks in cell-free systems. In Nucleic acids research 40 (8), pp. 3763–3774. DOI: 10.1093/nar/gkr1191.</p> | <p id="Karig2012">Karig, David K.; Iyer, Sukanya; Simpson, Michael L.; Doktycz, Mitchel J. (2012): Expression optimization and synthetic gene networks in cell-free systems. In Nucleic acids research 40 (8), pp. 3763–3774. DOI: 10.1093/nar/gkr1191.</p> | ||
| + | </div> | ||
| + | |||
| + | <div id="chromium" style="display: none"> | ||
<h1>Chromium</h1> | <h1>Chromium</h1> | ||
<!--We choose sfGFP as output signal for our sensors, because it’s measured more sensitive than RFP. For the <i>in vivo</i> measurement of our sensor system we cloned a devise that contains the chromium repressor protein ChrB and the chromium operator in front of our optimized UTR and sfGFP. | <!--We choose sfGFP as output signal for our sensors, because it’s measured more sensitive than RFP. For the <i>in vivo</i> measurement of our sensor system we cloned a devise that contains the chromium repressor protein ChrB and the chromium operator in front of our optimized UTR and sfGFP. | ||
| Line 78: | Line 206: | ||
--> | --> | ||
| − | + | <h2><i>in vivo</i></h2> | |
| − | <h2><i>in vivo</i></h2 | + | |
<p>The chromium sensor (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank"> BBa_K1758313 </a>) was constructed by using the basic construction we showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>. We work with the chromate inducible operon of <i>Ochrobactrum tritici</i> 5bvl1 which enables a resistance for chromium VI and superoxide. For our Sensor we used the Cr ( VI) dependent repressor chrB which was introduced by team BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank"> BBa_K1058007 </a>), and optimized this sequence for the use in <EM> E. coli </EM>. The associated chromium responsive promoter is ChrP (introduced by BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank"> BBa_K1058007 </a>). For output we used sfGFP and a 5’UTR untranslated region in front of sfGFP to optimize the expression of the reporter protein and increase its fluorescence.</p> | <p>The chromium sensor (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank"> BBa_K1758313 </a>) was constructed by using the basic construction we showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>. We work with the chromate inducible operon of <i>Ochrobactrum tritici</i> 5bvl1 which enables a resistance for chromium VI and superoxide. For our Sensor we used the Cr ( VI) dependent repressor chrB which was introduced by team BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank"> BBa_K1058007 </a>), and optimized this sequence for the use in <EM> E. coli </EM>. The associated chromium responsive promoter is ChrP (introduced by BIT 2013 (<a href="http://parts.igem.org/Part:BBa_K1758313" target="_blank"> BBa_K1058007 </a>). For output we used sfGFP and a 5’UTR untranslated region in front of sfGFP to optimize the expression of the reporter protein and increase its fluorescence.</p> | ||
| − | <p | + | <p>Our sensor for chromium detection consists of ChrB the repressor and the chromate specific promoter ChrP. The promoter is regulated by the ChrB, which binds Cr-ions. Behind the promoter is a sfGFP for detection of a fluorescence signal.</p> |
| − | <i>In vivo</i> we could show that the addition of different concentrations of chromium have different effects to transcription of sfGFP.</p | + | <i>In vivo</i> we could show that the addition of different concentrations of chromium have different effects to transcription of sfGFP.</p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| Line 96: | Line 223: | ||
</figure> | </figure> | ||
</br><p>Our data lead to the conclusion that in a cell based system it is possible to detect chromium. | </br><p>Our data lead to the conclusion that in a cell based system it is possible to detect chromium. | ||
| − | In contrast to our expectations with higher chromium concentrations we got lower fluorescence levels. These observations needed further investigation. </p | + | In contrast to our expectations with higher chromium concentrations we got lower fluorescence levels. These observations needed further investigation. </p> |
<h2><i>in vitro</i></h2> | <h2><i>in vitro</i></h2> | ||
| Line 125: | Line 252: | ||
<figcaption>Chromium sensor with alternative repressor build by team Dundee 2015, which has only the first 15 codons optimized in chromium specific cell extract under the induction withdifferent chromium concentrations. Error bars represent the standard deviation of three biological replicates.Data are normalised on chromiums influence to the cell extrat.</figcaption> | <figcaption>Chromium sensor with alternative repressor build by team Dundee 2015, which has only the first 15 codons optimized in chromium specific cell extract under the induction withdifferent chromium concentrations. Error bars represent the standard deviation of three biological replicates.Data are normalised on chromiums influence to the cell extrat.</figcaption> | ||
</figure> | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div id="copper" style="display: none"> | ||
<h1>Copper</h1> | <h1>Copper</h1> | ||
| − | <h2><i>in vivo</i></h2 | + | <h2><i>in vivo</i></h2> |
| − | <p>Our sensor for copper detection consists of CueR a MerR like activator and the copper specific promoter CopAP. The promoter is regulated by CueR, which binds Cu2+-ions. We also used a sfGFP behind the promoter for detection trough a fluorescence signal.</p> | + | <p>Our sensor for copper detection consists of CueR a MerR like activator and the copper specific promoter CopAP. The promoter is regulated by CueR, which binds Cu2+-ions. We also used a sfGFP behind the promoter for detection trough a fluorescence signal.</p> |
<p>For our copper sensor we used the native operator of cooper homeostasis from <i> E.coli </i> K12. And constructed a part (BBa_K1758324) which was constructed using the basic construction showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>.This operator includes the promoter (copAP), which is regulated by the repressor CueR. This is an important part for the aerobic copper tolerance. In BBa_K1758324 we combined the codon optimized CueR (<a href="http://parts.igem.org/Part:BBa_K1758320" target="_blank"> BBa_K1758320 </a>) under the control of a constitutive promoter with CopAP and sfGFP (<a href="http://parts.igem.org/Part:BBa_K1758321" target="_blank"> BBa_K1758321 </a>) for measuring output signals. Through the addition of a 5’UTR before the sfGFP we optimized the expression of sfGFP and increased fluorescence. </p> | <p>For our copper sensor we used the native operator of cooper homeostasis from <i> E.coli </i> K12. And constructed a part (BBa_K1758324) which was constructed using the basic construction showed in <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Project/HeavyMetals" target="_blank">Our biosensors</a>.This operator includes the promoter (copAP), which is regulated by the repressor CueR. This is an important part for the aerobic copper tolerance. In BBa_K1758324 we combined the codon optimized CueR (<a href="http://parts.igem.org/Part:BBa_K1758320" target="_blank"> BBa_K1758320 </a>) under the control of a constitutive promoter with CopAP and sfGFP (<a href="http://parts.igem.org/Part:BBa_K1758321" target="_blank"> BBa_K1758321 </a>) for measuring output signals. Through the addition of a 5’UTR before the sfGFP we optimized the expression of sfGFP and increased fluorescence. </p> | ||
| Line 138: | Line 268: | ||
<a href="https://static.igem.org/mediawiki/2015/9/90/Bielefeld-CeBiTec_Biolector_copper.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on OD600. Error bars represent the standard deviation of two biological replicates."><img src="https://static.igem.org/mediawiki/2015/9/90/Bielefeld-CeBiTec_Biolector_copper.jpg" alt="Adjusting the detection limit"></a> | <a href="https://static.igem.org/mediawiki/2015/9/90/Bielefeld-CeBiTec_Biolector_copper.jpg" data-lightbox="heavymetals" data-title="Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on OD600. Error bars represent the standard deviation of two biological replicates."><img src="https://static.igem.org/mediawiki/2015/9/90/Bielefeld-CeBiTec_Biolector_copper.jpg" alt="Adjusting the detection limit"></a> | ||
<figcaption>Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on OD600. Error bars represent the standard deviation of two biological replicates.</figcaption> | <figcaption>Time course of the induction of a copper biosensor with sfGFP for different copper concentrations in vivo. The data are measured with BioLector and normalized on OD600. Error bars represent the standard deviation of two biological replicates.</figcaption> | ||
| − | </figure | + | </figure> |
| − | <p><i>In vivo</i> we could show that the adding different concentrations of copper has effects on the transcription levels of sfGFP.</p | + | <p><i>In vivo</i> we could show that the adding different concentrations of copper has effects on the transcription levels of sfGFP.</p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="http://https://static.igem.org/mediawiki/2015/4/4e/Bielefeld-CeBiTec_Biolector_copper_Balkendiagramm.jpeg" data-lightbox="heavymetals" data-title="Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/4/4e/Bielefeld-CeBiTec_Biolector_copper_Balkendiagramm.jpeg" alt="Adjusting the detection limit"></a> | <a href="http://https://static.igem.org/mediawiki/2015/4/4e/Bielefeld-CeBiTec_Biolector_copper_Balkendiagramm.jpeg" data-lightbox="heavymetals" data-title="Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/4/4e/Bielefeld-CeBiTec_Biolector_copper_Balkendiagramm.jpeg" alt="Adjusting the detection limit"></a> | ||
<figcaption>Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes.</figcaption> | <figcaption>Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes.</figcaption> | ||
| − | </figure | + | </figure> |
| − | <p>The shown data suggest that sensing copper with our device is possible even if the detectable concentrations are higher than the desireble sensitivity limits. Therfore we tested the copper sensor in our <i>in vitro</i> transcription translation approach.</p | + | <p>The shown data suggest that sensing copper with our device is possible even if the detectable concentrations are higher than the desireble sensitivity limits. Therfore we tested the copper sensor in our <i>in vitro</i> transcription translation approach.</p> |
<h2><i>in vitro</i></h2> | <h2><i>in vitro</i></h2> | ||
| − | |||
In the following graphic the influences of different copper concentrations on the cell extact are shown | In the following graphic the influences of different copper concentrations on the cell extact are shown | ||
<!-- Einfluss von Kupfer auf den Zellextrakt, keinen negative Einfluss auf das CFPS so mit kann gezeigt werden dass dieses System relativ stabil gegenüber verschiedenen Kupferkonzentratione ist --> | <!-- Einfluss von Kupfer auf den Zellextrakt, keinen negative Einfluss auf das CFPS so mit kann gezeigt werden dass dieses System relativ stabil gegenüber verschiedenen Kupferkonzentratione ist --> | ||
| Line 157: | Line 286: | ||
<figcaption>Influence of different copper concentrations on our crude cell extract. Error bars represent the standard deviation of three biological replicates.</figcaption> | <figcaption>Influence of different copper concentrations on our crude cell extract. Error bars represent the standard deviation of three biological replicates.</figcaption> | ||
</figure></br> | </figure></br> | ||
| − | <p>As shown above copper has no negatice influence on the functuality of our cell extact. Therefore a ralatively stable system for copper sensing is provided.</p | + | <p>As shown above copper has no negatice influence on the functuality of our cell extact. Therefore a ralatively stable system for copper sensing is provided.</p> |
| − | <p>First tests with specific cell extract and different copper concentrations lead to further tests and normilisations.</p> | + | <p>First tests with specific cell extract and different copper concentrations lead to further tests and normilisations.</p> |
| − | + | ||
<!-- Induktion mit Kupfer im Kupfer spezifischen Extrakt --> | <!-- Induktion mit Kupfer im Kupfer spezifischen Extrakt --> | ||
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| Line 168: | Line 296: | ||
</figure> | </figure> | ||
| − | <p>Fluorescences normalised on coppers influence to the cell extract are shown above.<p/ | + | <p>Fluorescences normalised on coppers influence to the cell extract are shown above.<p/> |
<!--obrige Abbildung durch den errechneten Korrekturfaktor angepasst, da verschiedene Faktoren auf Zellextrakt wirken und so diesen beeinflussen.--> | <!--obrige Abbildung durch den errechneten Korrekturfaktor angepasst, da verschiedene Faktoren auf Zellextrakt wirken und so diesen beeinflussen.--> | ||
| Line 174: | Line 302: | ||
<a href="https://static.igem.org/mediawiki/2015/4/4c/Bielefeld-CeBiTec_correction_induction_copper_in_cueR_cell-extract.jpeg" data-lightbox="heavymetals" data-title="Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter without T7 in front of the operator site with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract."><img src="https://static.igem.org/mediawiki/2015/4/4c/Bielefeld-CeBiTec_correction_induction_copper_in_cueR_cell-extract.jpeg" alt="Adjusting the detection limit"></a> | <a href="https://static.igem.org/mediawiki/2015/4/4c/Bielefeld-CeBiTec_correction_induction_copper_in_cueR_cell-extract.jpeg" data-lightbox="heavymetals" data-title="Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter without T7 in front of the operator site with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract."><img src="https://static.igem.org/mediawiki/2015/4/4c/Bielefeld-CeBiTec_correction_induction_copper_in_cueR_cell-extract.jpeg" alt="Adjusting the detection limit"></a> | ||
<figcaption>Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter without T7 in front of the operator site with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract.</figcaption> | <figcaption>Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter without T7 in front of the operator site with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract.</figcaption> | ||
| − | </figure | + | </figure> |
| − | <p>In addition to the native promoter, operator device as measured above reporter constructs under the control of T7 promoter were tested.</p | + | <p>In addition to the native promoter, operator device as measured above reporter constructs under the control of T7 promoter were tested.</p> |
<!-- Es wurde auch das Konstrukt mit einen T7 davor eingesetzt, es zeichen sich unterschhiede inder Flurescens ausbeute, so mit ist für das CFPS system ein vorgeschalteter T7 sinnvoll zur besseren sensitivität des Systems. --> | <!-- Es wurde auch das Konstrukt mit einen T7 davor eingesetzt, es zeichen sich unterschhiede inder Flurescens ausbeute, so mit ist für das CFPS system ein vorgeschalteter T7 sinnvoll zur besseren sensitivität des Systems. --> | ||
| Line 181: | Line 309: | ||
<a href="https://static.igem.org/mediawiki/2015/c/ce/Bielefeld-CeBiTec_induction_T7-copAP_copper_in_cueR_cell-extract.jpeg" data-lightbox="heavymetals" data-title="Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction with different copper concentrations. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/c/ce/Bielefeld-CeBiTec_induction_T7-copAP_copper_in_cueR_cell-extract.jpeg" alt="Adjusting the detection limit"></a> | <a href="https://static.igem.org/mediawiki/2015/c/ce/Bielefeld-CeBiTec_induction_T7-copAP_copper_in_cueR_cell-extract.jpeg" data-lightbox="heavymetals" data-title="Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction with different copper concentrations. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/c/ce/Bielefeld-CeBiTec_induction_T7-copAP_copper_in_cueR_cell-extract.jpeg" alt="Adjusting the detection limit"></a> | ||
<figcaption>Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction with different copper concentrations. Error bars represent the standard deviation of three biological replicates.</figcaption> | <figcaption>Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction with different copper concentrations. Error bars represent the standard deviation of three biological replicates.</figcaption> | ||
| − | </figure | + | </figure> |
| − | <p>Compared to the former fluorecence leves the T7 reporter device showed higher levels therefore a reporter device under the control of T7 promoter is more suitable for our CFPS.</p | + | <p>Compared to the former fluorecence leves the T7 reporter device showed higher levels therefore a reporter device under the control of T7 promoter is more suitable for our CFPS.</p> |
| Line 190: | Line 318: | ||
<a href="https://static.igem.org/mediawiki/2015/0/01/Bielefeld-CeBiTec_correction_induction_T7-copAP_in_cueR_cell-extract.jpeg" data-lightbox="heavymetals" data-title="Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract.."><img src="https://static.igem.org/mediawiki/2015/0/01/Bielefeld-CeBiTec_correction_induction_T7-copAP_in_cueR_cell-extract.jpeg" alt="Adjusting the detection limit"></a> | <a href="https://static.igem.org/mediawiki/2015/0/01/Bielefeld-CeBiTec_correction_induction_T7-copAP_in_cueR_cell-extract.jpeg" data-lightbox="heavymetals" data-title="Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract.."><img src="https://static.igem.org/mediawiki/2015/0/01/Bielefeld-CeBiTec_correction_induction_T7-copAP_in_cueR_cell-extract.jpeg" alt="Adjusting the detection limit"></a> | ||
<figcaption>Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract.</figcaption> | <figcaption>Copper specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of copper inducible promoter with different copper concentrations. Error bars represent the standard deviation of three biological replicates. Data are normalized on coppers influence to the cell extract.</figcaption> | ||
| − | </figure | + | </figure> |
| − | <p> After normalising on coppers influcence to the cell extract these differecnces were even more obvious.</p></ | + | <p> After normalising on coppers influcence to the cell extract these differecnces were even more obvious.</p> |
| + | </div> | ||
| + | |||
| + | |||
| + | <div id="lead" style="display: none"> | ||
<h1>Lead</h1> | <h1>Lead</h1> | ||
<h2><i>in vivo</i></h2></br> | <h2><i>in vivo</i></h2></br> | ||
| − | <p>In addition to these we constructed a sensor for lead detection. It consists of PbrR, the repressor, and the lead specific promoter PbrA. The promoter is regulated by the RcnR, which binds Pb-ions. As the former sensors this one encloses a sfGFP for detection via fluorescence. </p | + | <p>In addition to these we constructed a sensor for lead detection. It consists of PbrR, the repressor, and the lead specific promoter PbrA. The promoter is regulated by the RcnR, which binds Pb-ions. As the former sensors this one encloses a sfGFP for detection via fluorescence. </p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| Line 209: | Line 341: | ||
</figure> | </figure> | ||
| − | <p> The differences between inductions with various lead concentrations are really slight therefore this sensor needs further optimization which was not possible in this limited time. But as there is a fluorescence response to lead this sensor has the potential work as expected. In the future a characterization in CFPS systems would be interesting. </p></ | + | <p> The differences between inductions with various lead concentrations are really slight therefore this sensor needs further optimization which was not possible in this limited time. But as there is a fluorescence response to lead this sensor has the potential work as expected. In the future a characterization in CFPS systems would be interesting. </p> |
| + | </div> | ||
| − | |||
| − | <h2><i>in vivo</i></h2 | + | <div id="mercury" style="display: none"> |
| + | <h1>Mercury</h1> | ||
| + | |||
| + | <h2><i>in vivo</i></h2> | ||
<!-- | <!-- | ||
| Line 229: | Line 364: | ||
</figure> | </figure> | ||
| − | <p><i>In vivo</i> data show a highly significant, well working sensor which even reacts to concentrations which are mentioned as drinking water guidelines by the WHO. <p | + | <p><i>In vivo</i> data show a highly significant, well working sensor which even reacts to concentrations which are mentioned as drinking water guidelines by the WHO. <p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
<a href="https://static.igem.org/mediawiki/2015/5/52/Bielefeld-CeBiTec_mercury_fluorescence_in_vivo_Balkendiagramm.jpeg" data-lightbox="heavymetals" data-title="Fluorescence levels at two different stages of cultivation. Shown are levels after 120 minutes and 190 minutes. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/5/52/Bielefeld-CeBiTec_mercury_fluorescence_in_vivo_Balkendiagramm.jpeg" alt="Adjusting the detection limit"></a> | <a href="https://static.igem.org/mediawiki/2015/5/52/Bielefeld-CeBiTec_mercury_fluorescence_in_vivo_Balkendiagramm.jpeg" data-lightbox="heavymetals" data-title="Fluorescence levels at two different stages of cultivation. Shown are levels after 120 minutes and 190 minutes. Error bars represent the standard deviation of three biological replicates."><img src="https://static.igem.org/mediawiki/2015/5/52/Bielefeld-CeBiTec_mercury_fluorescence_in_vivo_Balkendiagramm.jpeg" alt="Adjusting the detection limit"></a> | ||
<figcaption>Fluorescence levels at two different stages of cultivation. Shown are levels after 120 minutes and 190 minutes. Error bars represent the standard deviation of three biological replicates.</figcaption> | <figcaption>Fluorescence levels at two different stages of cultivation. Shown are levels after 120 minutes and 190 minutes. Error bars represent the standard deviation of three biological replicates.</figcaption> | ||
| − | </figure | + | </figure> |
| − | + | ||
| − | + | ||
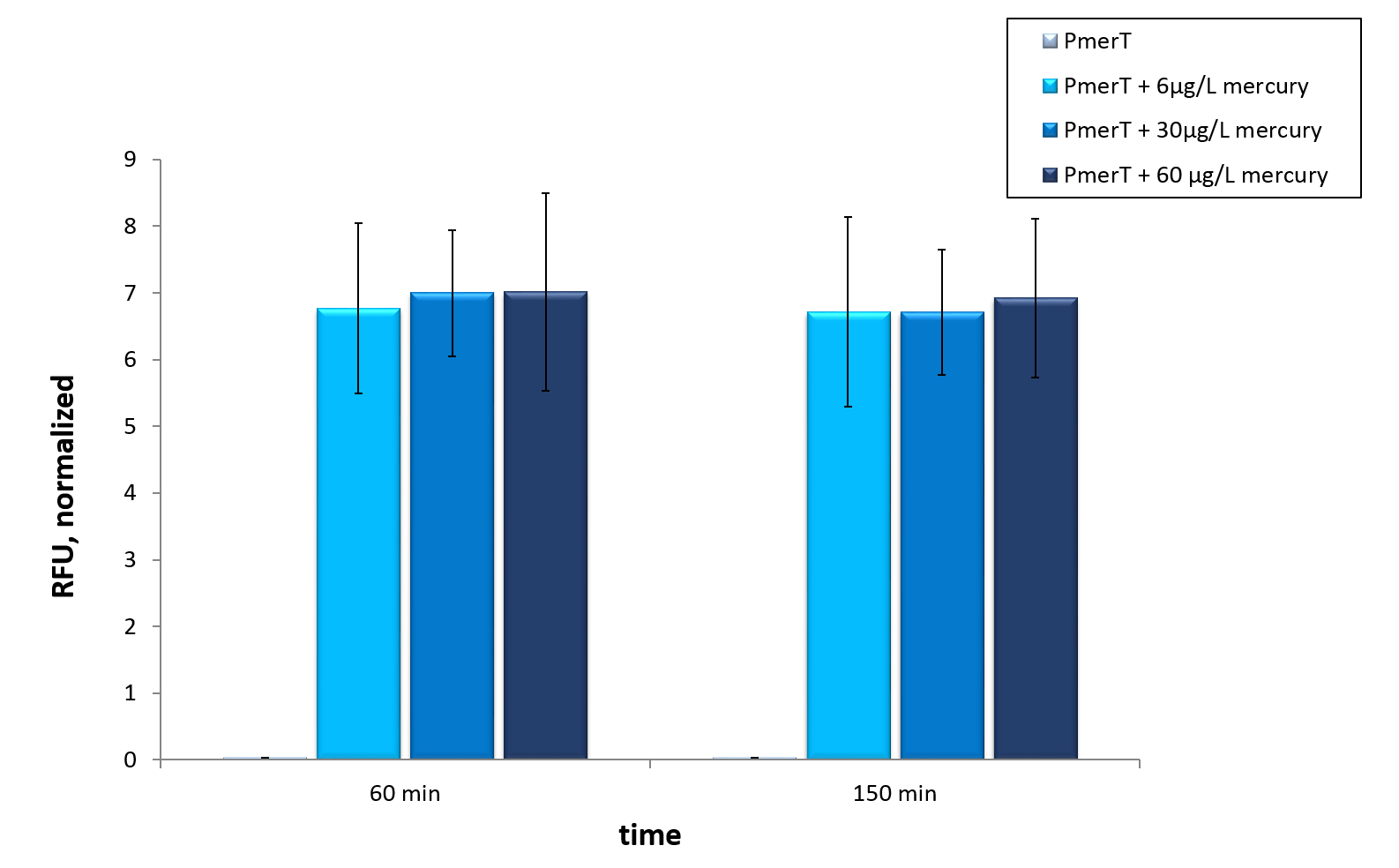
| + | <p>The mercury detection was measured during the cultivation of <i>E. coli</i> KRX at 37 °C. The strain contains the plasmid with the activator MerRunder the control of a constitutive promoter and the specific promoter with operator site which reacts to the activator with bound Hg-ions. The specific promoter is in front of sfGFP for measurment , so the mercury in the medium is detected directly.<i>In vivo</i> this sensor devise shows a fast answer to occurrence of his heavy metal contrary to the other sensor systems <i>in vivo</i>.</p> | ||
| − | <p> Therefore we tested our sensor <i>in vitro</i> to check if an already functioning highly optimized sensor provides required data for guideline detections </p | + | <p> Therefore we tested our sensor <i>in vitro</i> to check if an already functioning highly optimized sensor provides required data for guideline detections </p> |
| Line 263: | Line 397: | ||
<figcaption>Mercury specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of mercury inducible promoter without T7 in front of the operator site with different mercury concentrations. Error bars represent the standard deviation of three biological replicates.</figcaption> | <figcaption>Mercury specific cell extract made from <i>E. coli</i> cells which have already expressed the activator before cell extract production. Induction of mercury inducible promoter without T7 in front of the operator site with different mercury concentrations. Error bars represent the standard deviation of three biological replicates.</figcaption> | ||
</figure> | </figure> | ||
| + | </div> | ||
| + | |||
| + | <div id="nickel" style="display: none"> | ||
<h1>Nickel</h1> | <h1>Nickel</h1> | ||
| − | <h2><i>in vivo</i></h2 | + | <h2><i>in vivo</i></h2> |
| − | + | <p>In addition to these we aimed to construct a sensor for nickel detection. It consists of RcnR the repressor and the nickel specific promoter RcnA. The promoter is regulated by the RcnR, which binds Ni-ions. As the former sensors this one encloses a sfGFP for detection via Fluorescence.</p> | |
| − | <p> Our Nickel biosensor consists of parts of the rcn-operon from <i> E. coli </i> which codes for a nickel- and cobalt-efflux system. This system is highly sensitive to nickel. In absence of nickel or cobalt RcnR binds to the operator and inhibits the nickel responsive promoter. With Ni(II)-ions present the repression of the promoter RcnA will be reversed, because the repressor RcnR binds nickel-ions and cannot attach to the DNA. For our biosensor we construct the part (<a href="http://parts.igem.org/Part:BBa_K1758353" target="_blank"> BBa_K1758353 </a>by using the basic construction showed in <Our biosensors >. For this part we used the repressor RcnR under control of a constitutive promoter (<a href="http://parts.igem.org/Part:BBa_K1758350" target="_blank"> BBa_K1758350 </a>) and the nickel specific promoter RcnA with a 5’UTR in front of sfGFP (<a href="http://parts.igem.org/Part:BBa_K1758352" target="_blank"> BBa_K1758352 </a>) as reporter protein. </p> | + | <p>Our Nickel biosensor consists of parts of the rcn-operon from <i> E. coli </i> which codes for a nickel- and cobalt-efflux system. This system is highly sensitive to nickel. In absence of nickel or cobalt RcnR binds to the operator and inhibits the nickel responsive promoter. With Ni(II)-ions present the repression of the promoter RcnA will be reversed, because the repressor RcnR binds nickel-ions and cannot attach to the DNA. For our biosensor we construct the part (<a href="http://parts.igem.org/Part:BBa_K1758353" target="_blank"> BBa_K1758353 </a>by using the basic construction showed in <Our biosensors >. For this part we used the repressor RcnR under control of a constitutive promoter (<a href="http://parts.igem.org/Part:BBa_K1758350" target="_blank"> BBa_K1758350 </a>) and the nickel specific promoter RcnA with a 5’UTR in front of sfGFP (<a href="http://parts.igem.org/Part:BBa_K1758352" target="_blank"> BBa_K1758352 </a>) as reporter protein. </p> |
<figure style="width: 600px"> | <figure style="width: 600px"> | ||
| Line 281: | Line 418: | ||
<figcaption>Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes. Error bars represent the standard deviation of three biological replicates.</figcaption> | <figcaption>Fluorescence levels at three different stages of cultivation. Shown are levels after 60 minutes, 150 minutes and 650 minutes. Error bars represent the standard deviation of three biological replicates.</figcaption> | ||
</figure> | </figure> | ||
| − | <p> With this sensor no production of sfGFp via fluorescence level change could be detected. Therefore this sensor is not suitable for approach. Therefore no <i> in vitro </i> data using CFPS were taken.</p></ | + | <p> With this sensor no production of sfGFp via fluorescence level change could be detected. Therefore this sensor is not suitable for approach. Therefore no <i> in vitro </i> data using CFPS were taken.</p> |
| − | + | </div> | |
</div> | </div> | ||
| Line 288: | Line 425: | ||
<div class="row footer"> | <div class="row footer"> | ||
<div class="col-md-2 col-md-offset-10 text-center"> | <div class="col-md-2 col-md-offset-10 text-center"> | ||
| − | <a type="button" class="btn btn-default btn-next" href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Results/DateRapeDrugs"><img src="https://static.igem.org/mediawiki/2015/5/51/Bielefeld-CeBiTec_iGEM_logo.png"><p>And the date rape drugs?</p></a> | + | <a type="button" class="btn btn-default btn-next" href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Results/DateRapeDrugs"><img src="https://static.igem.org/mediawiki/2015/5/51/Bielefeld-CeBiTec_iGEM_logo.png"><p>And what about the date rape drugs?</p></a> |
</div> | </div> | ||
</div> | </div> | ||
Revision as of 20:55, 15 September 2015
Heavy Metals
Zusammenfassung in ganz wenigen Worten.
The different sensors we worked with were characterized in vivo as well as in vitro.
We tested the influence of each heavy metal on our sensors in vivo Therefore we used heavy metal concentrations based on heavy metal occurrences measured all over the world.