Difference between revisions of "Team:UESTC Software/Practices"
| (32 intermediate revisions by 3 users not shown) | |||
| Line 14: | Line 14: | ||
<link rel="stylesheet" href="https://2015.igem.org/Template:UESTC_Software/divide_dou_css?action=raw&ctype=text/css">//我的分页 | <link rel="stylesheet" href="https://2015.igem.org/Template:UESTC_Software/divide_dou_css?action=raw&ctype=text/css">//我的分页 | ||
<script src="https://2015.igem.org/Template:UESTC_Software/headjs?action=raw&ctype=text/javascript"></script> | <script src="https://2015.igem.org/Template:UESTC_Software/headjs?action=raw&ctype=text/javascript"></script> | ||
| − | + | <!--<script src="https://2015.igem.org/Template:UESTC_Software/jquery_nav_js?action=raw&ctype=text/javascript"></script>--> | |
| − | + | <script type="text/javascript"> | |
| + | /*$(document).ready(function(){ | ||
| + | $(".dou_navbar ul").onePageNav(); | ||
| + | })*/ | ||
| + | </script> | ||
<header class="dou_header"> | <header class="dou_header"> | ||
<div class="header_logo"> | <div class="header_logo"> | ||
| Line 29: | Line 33: | ||
<a class="" href="javascript:void(0)">PROJECT<br/><i class="iconfont"></i></a> | <a class="" href="javascript:void(0)">PROJECT<br/><i class="iconfont"></i></a> | ||
<ul id="menu_pro" class="dou_divide_nav_second clearfix"> | <ul id="menu_pro" class="dou_divide_nav_second clearfix"> | ||
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/Description | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Description">Description</a></li> |
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/ | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Design">Design</a></li> |
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/ | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Software">Software</a></li> |
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/Modeling | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Modeling">Modeling</a></li> |
| + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Validation">Validation</a></li> | ||
| + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Results">Results</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
<li> | <li> | ||
| − | <a href="javascript:void(0)"> | + | <a href="javascript:void(0)">DOCUMENTS<br/><i class="iconfont"></i></a> |
| − | <ul id=" | + | <ul id="menu_req" class="dou_divide_nav_second"> |
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/ | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/User_guide">User Guide</a></li> |
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/ | + | <li style=""><a href="https://2015.igem.org/Team:UESTC_Software/Documentation">API</a></li> |
| + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Notebook">Notebook</a></li> | ||
</ul> | </ul> | ||
| − | |||
| − | |||
| − | |||
</li> | </li> | ||
<li class="choose"> | <li class="choose"> | ||
| − | + | <a href="https://2015.igem.org/Team:UESTC_Software/Practices">HUMAN<br/>PRACTICE</a> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</li> | </li> | ||
<li> | <li> | ||
<a href="javascript:void(0)">REQUIREMENTS<br/><i class="iconfont"></i></a> | <a href="javascript:void(0)">REQUIREMENTS<br/><i class="iconfont"></i></a> | ||
<ul id="menu_req" class="dou_divide_nav_second"> | <ul id="menu_req" class="dou_divide_nav_second"> | ||
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/Medals | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Medals">Medals</a></li> |
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/Safety | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Safety">Safety</a></li> |
| − | <li><a href="https://2015.igem.org/Team:UESTC_Software/ | + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Collaborations">Collaborations</a></li> |
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="javascript:void(0)">TEAM<br/><i class="iconfont"></i></a> | ||
| + | <ul id="menu_team" class="dou_divide_nav_second"> | ||
| + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Members">Members</a></li> | ||
| + | <li><a href="https://2015.igem.org/Team:UESTC_Software/Attributions">Attributions</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 70: | Line 77: | ||
</div> | </div> | ||
<section class="cd_half_section" id="c_h_s_meet"> | <section class="cd_half_section" id="c_h_s_meet"> | ||
| − | <h1> | + | <h1>Human Practice</h1> |
| + | <p>Photo From © PEXELS</p> | ||
</section> | </section> | ||
| + | <!--<div class="dou_navbar clearfix"> | ||
| + | <div class="dou_navbar_wrap"> | ||
| + | <ul> | ||
| + | <li class="dou_one"><a class=" current" href="#understand">Understand</a></li> | ||
| + | <li class="dou_second"><a href="#investigate">Investigate</a> | ||
| + | <li class="second_nav"><a href="#investigate">Meet-up</a></li> | ||
| + | <li class="second_nav"><a href="#hku">Communication</a></li> | ||
| + | <li class="second_nav"><a href="#game">Game</a></li> | ||
| + | <li class="second_nav"><a href="#invest_results">Results</a></li> | ||
| + | <li class="dou_one"><a href="#conclusion">Conclusion</a></li> | ||
| + | |||
| + | </ul> | ||
| + | <div class="dou_navButton"> | ||
| + | < | ||
| + | </div> | ||
| + | </div> | ||
| + | </div>--> | ||
<section class="cd-section"> | <section class="cd-section"> | ||
<div class="cd-main-content"> | <div class="cd-main-content"> | ||
<div class="dou_m_c_p"> | <div class="dou_m_c_p"> | ||
<div class="dou_title"> | <div class="dou_title"> | ||
| − | <h2> | + | <h2 id="understand">How do we understand our human practice?</h2> |
</div> | </div> | ||
<p> | <p> | ||
| − | + | We are a software team which is not involved in any experiments. So the sustainability of software is the most notable issue, rather than ethics or social safety. After investigating and discussion, we found two ways to solve this problem. | |
| − | </p | + | </p> |
<p> | <p> | ||
| − | + | √Finding science research groups or enterprises that show great interest in our software in order to get their help to continue our project development. | |
| − | </p | + | </p> |
<p> | <p> | ||
| − | + | √Cooperate with people who are interested in our project. Voluntary efforts by these people will help improve and continue our project. | |
</p> | </p> | ||
</div> | </div> | ||
| − | <div class="dou_m_c_pic"> | + | <!--<div class="dou_m_c_pic"> |
<div class="dou_m_c_pic_wrap real_pic_wrap clearfix"> | <div class="dou_m_c_pic_wrap real_pic_wrap clearfix"> | ||
| − | <img id="real_pic1" src="/ | + | <img id="real_pic1" src="assets/human_practice1.png"/> |
| − | + | <img id="real_pic2" src="assets/human_practice1.png"/> | |
| − | + | <img id="real_pic3" src="assets/human_practice1.png"/> | |
| + | |||
</div> | </div> | ||
| − | </div> | + | </div>--> |
</div> | </div> | ||
| Line 103: | Line 129: | ||
<div class="dou_m_c_p"> | <div class="dou_m_c_p"> | ||
<div class="dou_title"> | <div class="dou_title"> | ||
| − | <h2> | + | <h2>Investigate and solve the problem</h2> |
| + | <h3 id="investigate">① We hold the iGEM Southwest Union of China</h3> | ||
</div> | </div> | ||
| − | <p | + | <p> |
| − | + | We found an iGEM organization for academic communication, which promotes the ideas exchanging of iGEM project and synthetic biology with students from other universities in southwest of China this year. On July 25, 2015, we invited Sichuan University , Sichuan Agricultural University, Southwest University of Nationalities,Chengdu Shude High School and Bio maker to participate in the party which is organized by us(UESTC). | |
| + | </p> | ||
| + | <p> | ||
| + | Firstly, our instructor Huang Jian introduced the background, some components of iGEM and several precautions according to his experience in the communication meeting. Then, each school had its own report. Sichuan University iGEM experimental team introduced their own nitrogen and carbon sequestration technologies in microorganism. They also met a problem on SD sequence finding which influences the development of their project. However, we studied how to find out SD sequence in DNA sequence and analyze the strength of the SD sequence that affects translation issues to help them solve their problem. (For more information, go to the collaboration page) Another experimental team of University of Electronic Science and Technology of China shared their novel magneto tactic bacterium. What’s more, we not only shared our stories, presented the software development but also had a heated and intensive discussion on synthetic biology with students who were present. With the idea-exchanging more and more extensive, students from different universities gradually obtained a further and clearer understanding of synthetic biology, especially the minimal cell, the minimal gene set and the relationship between them. At the same time, our introduction made everybody get through to our project and promoted much more interest. This is a good start for the collaboration of iGEM teams in southwest of China. In the end, our instructor Guo Fengbiao made a summary and encouraged us to do what we lobe. After the seminar, we wrote press releases and published in school website of UESTC, attracting more people and making them know much more information about iGEM. | ||
| + | |||
| + | |||
</p> | </p> | ||
</div> | </div> | ||
<div class="dou_m_c_pic"> | <div class="dou_m_c_pic"> | ||
<div class="dou_m_c_pic_wrap clearfix"> | <div class="dou_m_c_pic_wrap clearfix"> | ||
| − | <img src="/wiki/images/ | + | <img src="/wiki/images/3/3d/Uestc_software_Human_practice1.png"/><!--/wiki/images/3/3d/Uestc_software_Human_practice1.png--> |
</div> | </div> | ||
</div> | </div> | ||
| + | |||
</div> | </div> | ||
| Line 121: | Line 154: | ||
<div class="dou_m_c_p"> | <div class="dou_m_c_p"> | ||
<div class="dou_title"> | <div class="dou_title"> | ||
| − | + | <h3 id="hku">② Communication with HKU and senior high school</h3> | |
| − | < | + | |
</div> | </div> | ||
<p> | <p> | ||
| − | + | On June 20, 2015, Faculty of Medicine came to our school to introduce their genome sequencing for dissecting transmission pathways of Elizabethkingia anopheles, whose data was very sophisticated and enormous. But their research was charming and provided our project with some advice as well. | |
| − | </p>< | + | </p> |
| + | <div class="dou_m_c_pic"> | ||
| + | <div class="dou_m_c_pic_wrap clearfix"> | ||
| + | <img src="/wiki/images/9/93/Uestc_software_human_practice4.png"/><!--/wiki/images/c/c8/Uestc_software_Human_practice2.png--> | ||
| + | </div> | ||
| + | </div> | ||
| + | <p> | ||
| + | The next day, we came to Chengdu NO.7 High School JiaXiang Foreign Language School to preach iGEM competition and related knowledge on synthetic biology with the students from Faculty of Engineering of HKU. We together introduced our software to the high school students, which was a very interesting and significance experience to both of us. Fortunately, the senior high school students showed great interest in synthetic biology and our software, which inspiring us greatly. Except presenting our own iGEM work and sharing a lot of experience, we would also have more communication in the future. | ||
| + | </p> | ||
</div> | </div> | ||
| − | + | <div class="dou_m_c_pic"> | |
<div class="dou_m_c_pic_wrap clearfix"> | <div class="dou_m_c_pic_wrap clearfix"> | ||
| − | <img src="/wiki/images/ | + | <img src="/wiki/images/c/c8/Uestc_software_Human_practice2.png"/><!--/wiki/images/c/c8/Uestc_software_Human_practice2.png--> |
</div> | </div> | ||
</div> | </div> | ||
| + | |||
</div> | </div> | ||
| + | </section> | ||
| + | <section class="cd-section"> | ||
| + | <div class="cd-main-content"> | ||
| + | <div class="dou_m_c_p"> | ||
| + | <div class="dou_title"> | ||
| + | <h3 id="game">③ Game--Father’s Gene Detector</h3> | ||
| + | </div> | ||
| + | <p> | ||
| + | We developed an interesting game based on the strategy of our software, which could make users a complete understanding of our half-retaining method. Actually, it is a game for simulating minimal genome selection and linking with PC—side software, developed completely by Xcode. With careful preparation of our art designers, we use five roles instead of nine roles in ancient story, which could reduce the difficulty of the game to some extent. Generally, it will let players enjoy fun and the feelings of freshness of ancient story. Through the game, we wish more and more people experience the function of essential gene visually as well as known more about our software and synthetic biology. | ||
| + | </p> | ||
| + | <div class="dou_m_c_pic"> | ||
| + | <div class="dou_m_c_pic_wrap clearfix"> | ||
| + | <img style="width:24%" src="/wiki/images/b/bc/Uestc_software_human_practice3.png"/> | ||
| + | <img style="width:24%" src="/wiki/images/9/98/Uestc_software_human_practice3_2.png"/> | ||
| + | <img style="width:24%" src="/wiki/images/d/d6/Uestc_software_human_practice3_3.png"/> | ||
| + | <img style="width:24%" src="/wiki/images/c/c4/Uestc_software_human_practice3_4.png"/> | ||
| + | </div> | ||
| + | </div> | ||
| + | <p>In order to make the comprehension of game by people without the background of biology become easier, we also invited some foreign students to play our game after the publication of version 1.0. It is interesting to note that most of them are attracted by it. Meanwhile,we also collect some comments from these student ,which is more objective. | ||
| + | </p> | ||
| + | <div class="dou_m_c_pic"> | ||
| + | <div class="dou_m_c_pic_wrap clearfix"> | ||
| + | <img src="/wiki/images/4/4a/Uestc_software_human_practice6.png"/> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </section> | ||
| + | <section class="cd-section"> | ||
| + | <div class="cd-main-content"> | ||
| + | <div class="dou_m_c_p"> | ||
| + | <div class="dou_title"> | ||
| + | <h3 id="invest_results">④ Investigation Results</h3> | ||
| + | </div> | ||
| + | <div class="dou_m_c_pic"> | ||
| + | <div class="dou_m_c_pic_wrap clearfix"> | ||
| + | <img style="width:80%;" src="/wiki/images/6/63/Uestc_software_human_practice5.png"/> | ||
| + | </div> | ||
| + | </div> | ||
| + | <p> | ||
| + | This year, our team UESTC-Software has conducted many human practices to disseminate iGEM and our project. One of them is a questionnaire that gathers the information of public awareness of knowledge on synthetic biology and our software. The pie chart shows the proportion of number of different students and social elites who are participate in our human practices. | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </section> | ||
| + | <section class="cd-section"> | ||
| + | <div class="cd-main-content"> | ||
| + | <div class="dou_m_c_p"> | ||
| + | <div class="dou_title"> | ||
| + | <h2 id="conclusion">Conclusion</h2> | ||
| + | </div> | ||
| + | <p> | ||
| + | As a result of our hard work and research, it’s very fortunate that our software is attractive enough to draw the professors and entrepreneurs. In addition, through our efforts, our software has attracted many students majored in Bioinformatics and Qt developers as well, for the reason that we follow the basic principle of KISS (keep it simple, Stupid). | ||
| + | </p> | ||
| + | <p> | ||
| + | In the future, we will pay much more attention to coding standardization and document perfection. We also wish there are more and more people reuse our software, join us and continue our work. | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
</section> | </section> | ||
Latest revision as of 16:49, 18 September 2015
Human Practice
Photo From © PEXELS
How do we understand our human practice?
We are a software team which is not involved in any experiments. So the sustainability of software is the most notable issue, rather than ethics or social safety. After investigating and discussion, we found two ways to solve this problem.
√Finding science research groups or enterprises that show great interest in our software in order to get their help to continue our project development.
√Cooperate with people who are interested in our project. Voluntary efforts by these people will help improve and continue our project.
Investigate and solve the problem
① We hold the iGEM Southwest Union of China
We found an iGEM organization for academic communication, which promotes the ideas exchanging of iGEM project and synthetic biology with students from other universities in southwest of China this year. On July 25, 2015, we invited Sichuan University , Sichuan Agricultural University, Southwest University of Nationalities,Chengdu Shude High School and Bio maker to participate in the party which is organized by us(UESTC).
Firstly, our instructor Huang Jian introduced the background, some components of iGEM and several precautions according to his experience in the communication meeting. Then, each school had its own report. Sichuan University iGEM experimental team introduced their own nitrogen and carbon sequestration technologies in microorganism. They also met a problem on SD sequence finding which influences the development of their project. However, we studied how to find out SD sequence in DNA sequence and analyze the strength of the SD sequence that affects translation issues to help them solve their problem. (For more information, go to the collaboration page) Another experimental team of University of Electronic Science and Technology of China shared their novel magneto tactic bacterium. What’s more, we not only shared our stories, presented the software development but also had a heated and intensive discussion on synthetic biology with students who were present. With the idea-exchanging more and more extensive, students from different universities gradually obtained a further and clearer understanding of synthetic biology, especially the minimal cell, the minimal gene set and the relationship between them. At the same time, our introduction made everybody get through to our project and promoted much more interest. This is a good start for the collaboration of iGEM teams in southwest of China. In the end, our instructor Guo Fengbiao made a summary and encouraged us to do what we lobe. After the seminar, we wrote press releases and published in school website of UESTC, attracting more people and making them know much more information about iGEM.

② Communication with HKU and senior high school
On June 20, 2015, Faculty of Medicine came to our school to introduce their genome sequencing for dissecting transmission pathways of Elizabethkingia anopheles, whose data was very sophisticated and enormous. But their research was charming and provided our project with some advice as well.

The next day, we came to Chengdu NO.7 High School JiaXiang Foreign Language School to preach iGEM competition and related knowledge on synthetic biology with the students from Faculty of Engineering of HKU. We together introduced our software to the high school students, which was a very interesting and significance experience to both of us. Fortunately, the senior high school students showed great interest in synthetic biology and our software, which inspiring us greatly. Except presenting our own iGEM work and sharing a lot of experience, we would also have more communication in the future.

③ Game--Father’s Gene Detector
We developed an interesting game based on the strategy of our software, which could make users a complete understanding of our half-retaining method. Actually, it is a game for simulating minimal genome selection and linking with PC—side software, developed completely by Xcode. With careful preparation of our art designers, we use five roles instead of nine roles in ancient story, which could reduce the difficulty of the game to some extent. Generally, it will let players enjoy fun and the feelings of freshness of ancient story. Through the game, we wish more and more people experience the function of essential gene visually as well as known more about our software and synthetic biology.




In order to make the comprehension of game by people without the background of biology become easier, we also invited some foreign students to play our game after the publication of version 1.0. It is interesting to note that most of them are attracted by it. Meanwhile,we also collect some comments from these student ,which is more objective.

④ Investigation Results
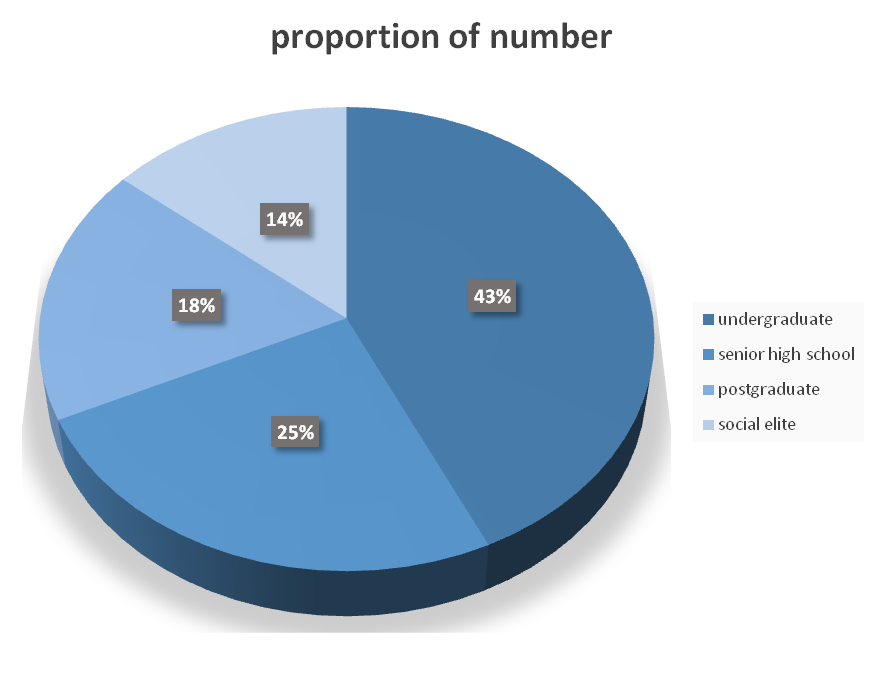
This year, our team UESTC-Software has conducted many human practices to disseminate iGEM and our project. One of them is a questionnaire that gathers the information of public awareness of knowledge on synthetic biology and our software. The pie chart shows the proportion of number of different students and social elites who are participate in our human practices.
Conclusion
As a result of our hard work and research, it’s very fortunate that our software is attractive enough to draw the professors and entrepreneurs. In addition, through our efforts, our software has attracted many students majored in Bioinformatics and Qt developers as well, for the reason that we follow the basic principle of KISS (keep it simple, Stupid).
In the future, we will pay much more attention to coding standardization and document perfection. We also wish there are more and more people reuse our software, join us and continue our work.
