Difference between revisions of "Team:UCLA/Notebook/Spider Silk Genetics/2 September 2015"
Vinsonclam (Talk | contribs) |
Vinsonclam (Talk | contribs) |
||
| Line 80: | Line 80: | ||
| hold | | hold | ||
|} | |} | ||
| + | *Verified 5 uL of each reaction on 1% TAE gel with 2 uL of 1 kb ladder. | ||
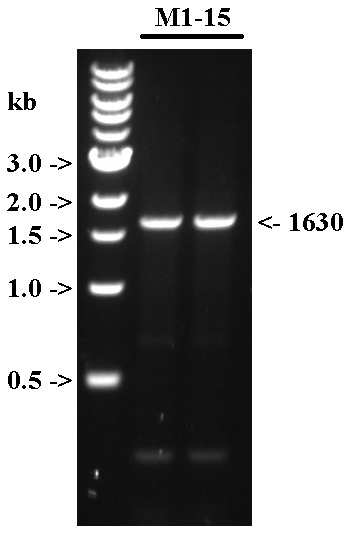
| + | [[File:9 2 2015 UCLA ICA.jpg|none|thumb|500px|'''Fig. 1''' PCR amplification of M1-15 using previous M1-15 as template. The expected band size is 1630 bp.]] | ||
| + | *Decided that the band looks clean enough, so will PCR purify and not gel purify. | ||
| + | **Eluted in 12 uL. | ||
| + | |||
| + | ==Digestion for M1-15== | ||
| + | *Digest 5 uL of the PCR product with E,P and 5 uL of PCR product with X,P. | ||
| + | *Digest in a 50 uL reaction with buffer 2.1 and 1 uL of each relevant enzyme. | ||
| + | *Digest at 37 C for 1.5 hrs, heat kill at 65 C for 20 min. | ||
| + | *PCR purified the digestion product. | ||
| + | |||
| + | ==Ligation for M1-15== | ||
| + | *Ligate E,P digested products into pSB1C3. | ||
| + | *Ligate X,P digested products into BBa_K525998(S,P) | ||
| + | *Ligate at a 1:3 molar vector to insert ratio. | ||
| + | *Ligate at 25 C for 1 hr, heat kill at 65 C for 20 min. | ||
| + | |||
| + | ==Transformation== | ||
| + | *Transform M1-15(1C3) into DH5(alpha) electrocompetent cells. | ||
| + | **Dialyze against ultra-pure ddH2O. | ||
| + | **Tried 3 times. First two times, the sample arced. Third time transformed 2 uL of a a 1:50 idlution. | ||
| + | **Arc timme 5.4 s. | ||
| + | *Transform M1-15(T7) into BL21(DE3) chemically competent cells. | ||
Latest revision as of 21:36, 6 September 2015
Contents
9/2/2015
Sequencing Results
- M1-12(1C3)
- 1: GOOD
- 2: Bad, deletion in suffix
- 3: GOOD
- M1-12(T7)
- 1: incorrect sequence
- 2: GOOD
- 3: Bad, no reverse priming
- M1/2[1:2]-12(1C3)
- 1: GOOD
- 2: GOOD
- 3: GOOD
- M1/2[1:2]-12(T7)
- 1: Bad, no priming
- 2: Bad, non-specific
- 3: GOOD
Gel Purification for M1-15
- Used zymo kit.
- Yield 31.64 ng/uL A: 2.47
- Need to do another PCR to get more.
PCR Amplification for M1-15
- Used 1:1000 dilution of M1-15 from above as template.
- Used ~100 pg of template in reaction.
- Set up 2x 50 uL reactions
| Volume (uL) | |
|---|---|
| 5x Q5 Buffer | 10 |
| 10 mM dNTPs | 1 |
| 10 uM For (F-03) | 2.5 |
| 10 uM Rev (G-03) | 2.5 |
| Template (~30 pg/uL) | 3j |
| Q5 Polymerase | 0.5 |
| ddH2O | 30.5 |
| Total | 50 |
| 98 C | 30 sec |
| 98 C | 10 sec |
| 66 C | 20 sec |
| 72 C | 40 sec |
| repeat from step 2 | 25x |
| 72 C | 2 min |
| 12 C | hold |
- Verified 5 uL of each reaction on 1% TAE gel with 2 uL of 1 kb ladder.
- Decided that the band looks clean enough, so will PCR purify and not gel purify.
- Eluted in 12 uL.
Digestion for M1-15
- Digest 5 uL of the PCR product with E,P and 5 uL of PCR product with X,P.
- Digest in a 50 uL reaction with buffer 2.1 and 1 uL of each relevant enzyme.
- Digest at 37 C for 1.5 hrs, heat kill at 65 C for 20 min.
- PCR purified the digestion product.
Ligation for M1-15
- Ligate E,P digested products into pSB1C3.
- Ligate X,P digested products into BBa_K525998(S,P)
- Ligate at a 1:3 molar vector to insert ratio.
- Ligate at 25 C for 1 hr, heat kill at 65 C for 20 min.
Transformation
- Transform M1-15(1C3) into DH5(alpha) electrocompetent cells.
- Dialyze against ultra-pure ddH2O.
- Tried 3 times. First two times, the sample arced. Third time transformed 2 uL of a a 1:50 idlution.
- Arc timme 5.4 s.
- Transform M1-15(T7) into BL21(DE3) chemically competent cells.