Team:ETH Zurich/Results
- Project
- Modeling
- Lab
- Human
Practices - Parts
- About Us
Results
We are excited to present a lot of meaningful results, including the characterization of parts of the natural E. coli lldPRD operon on which there is only a very limited amount of information present in the literature up to date. In addition, we attempted to reproduce previously published results on the apoptosis induction in mammalian cancer cell lines using sTRAIL, and tried to validate the Warburg effect for the same cell lines.
For a better understanding of the results of our project, we divided this part into four parts:
Experiments involving mammalian cells
Our first step needed to check the viability of our experimental set-up was to confirm the data found in the literature regarding the sensitivity to TRAIL [Thomas, 2008] and the elevated lactate production observed in cancer cells, but not in normal cells.
Sensitivity to TRAIL
Background
Apoptosis can be induced through a protein called TNF-related Apoptosis Inducing Ligand (TRAIL). TRAIL is a transmembrane protein which can be found also in solubilised form [de Bruyne et al, 2013 ] that recognizes the death receptors DR4, DR5, DCR1, and DCR2 [Johnstone et al, 2008] and it has been used in cancer treatment inducing the apoptosis of cancerous cells but with low cytotoxicity for healthy ones [Johnstone et al, 2008; Zhang et al, 2005].
Experiment overview
The first signal in our system is based on making cancer cells apoptotic while healthy cells remain untouched. Due to the properties of TRAIL explained before, we used it for a selective apoptosis induction in cancer cells.
The aim of this experiment was to check if cancer cells effectively undergo apoptosis after .ong the incubation should last.
To test the predisposition of cancer cells to TRAIL-induced apoptosis we chose Jurkat cells, which are an immortalized human T lymphocyte cell line; and HL-60 cells, a line derived from acute myeloid leukemia. For a control with healthy cells we used a murine fibroblast cell line (3T3).
Methods and Results
Our first experiment consisted in incubating Jurkat and 3T3 cells with TRAIL for 4h and 6h (see protocol about apoptosis sensibility). Concentrations of 0, 50 and 100 ng/mL of TRAIL were used with each cell line. A positive control for Jurkat cells was applied by incubating these cells with PMA and ionomycin overnight. After that, FACS analysis was performed. To differentiate between apoptosis and necrosis, PI dye was used to stain necrotic cells. Apoptotic cells were detected by using Annexin V labelled with Alexa 488 (Life Technologies).
Unfortunately, despite doing several trials in which we incremented the number of hours for incubation (up to 48h) and tried different concentrations of TRAIL, we were not able to replicate the results we found in the literature. To see where the problem was, we decided to incubate the cells with 1000 ng/mL of TRAIL over a period of 48 h. If TRAIL was functional, we would expect to see the cells with a different morphology, which would mean that our detection system is not working even though apoptosis was properly induced. If TRAIL was not functional, we expect not to see morphological differences. After the 48h we saw that there were no differences in the morphology of the cells, which lead us to suppose that probably the sTRAIL we bought was not working as expected.
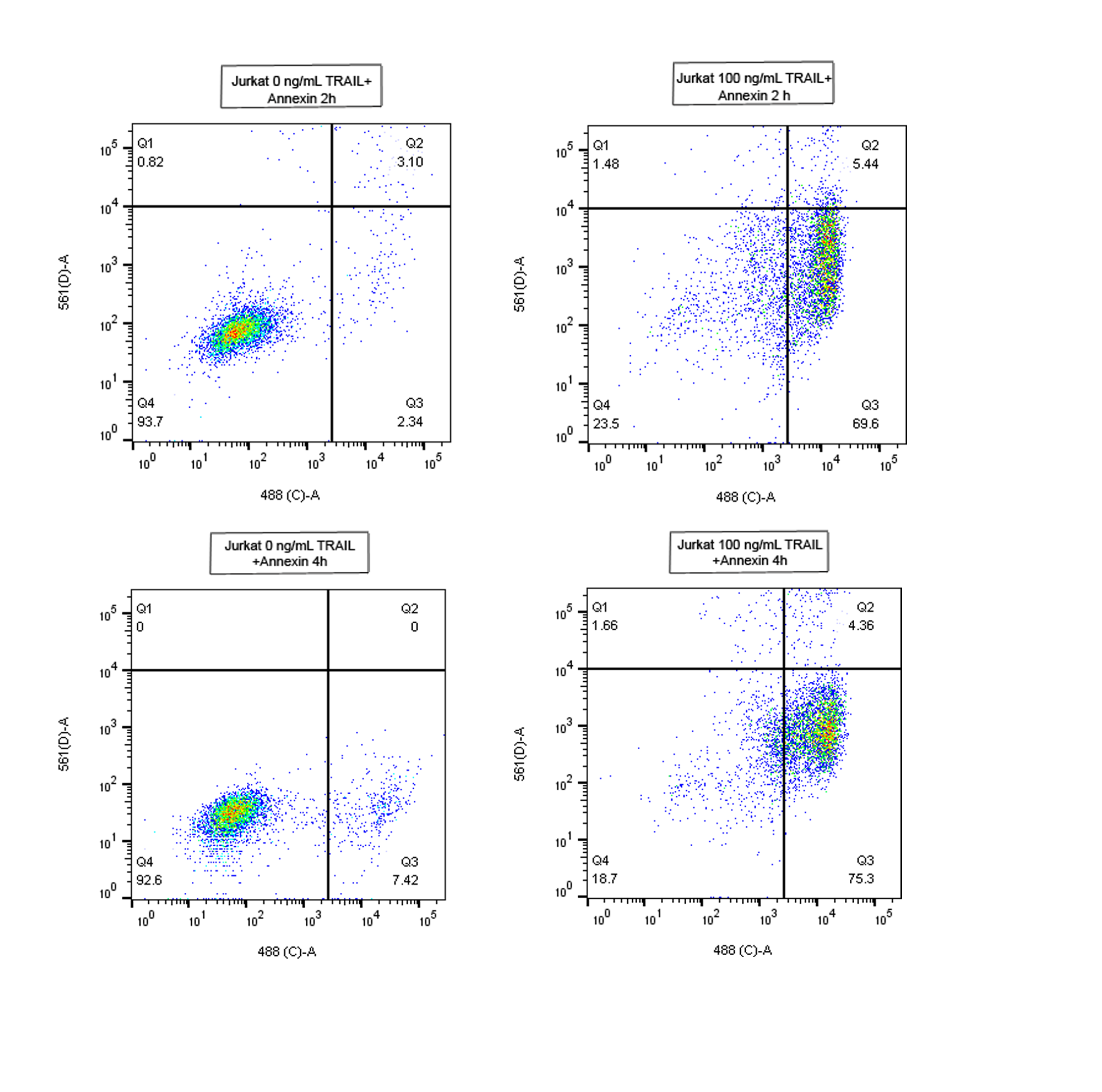
Our next step was to purchase another TRAIL called killerTRAIL (Enzo). To check whether this time our experiments were working, we added 100 ng/mL of TRAIL to Jurkat cells and measured the apoptosis at 2 h and 4 h. We observed that in 2 h apoptosis increased from about 2% of the population in non-treated samples to almost 70% in cells incubated with TRAIL. After 4 h the apoptotic cell number increased up to 75% (see Figure 1). As the number of apoptotic cells is increasing slowly after 2 h, we decided to use this as our standard time for TRAIL incubation.
Once we confirmed that TRAIL was effectively inducing apoptosis in Jurkat cells, we wanted todo the complete experiment, in which we determine the concentration of TRAIL we will use in future experiments and contrast Jurkat cells with 3T3 (a non-cancerous cell line).
Jurkat cells and 3T3 were incubated for 2 h in 0, 10, 50 and 100 ng/mL of TRAIL. Then, we followed the protocol detailed in Experiments. Annexin V was added to detect apoptosis, as it binds to phosphatidylserine which is found in higher proportion in apoptotic cells. To differentiate apoptotic and necrotic cells, PI dye was added. Annexin V and PI were detected by an emission filter at 488 and 561 nm, respectively.
Our data shows that 3T3 are not affected by TRAIL after 2h, whereas Jurkat cells are (as have been shown in the previous experiment). For Jurkat cells we decided to use a concentration of 100 ng/mL, as it allows to have a higher number of apoptotic cells in a relatively short amount of time (2 h).


Figure 3. 3T3 (upper image) and Jurkat cells (lower image) with 0, 10, 50 and 100 ng/mL of TRAIL after 2 h incubation. Annexin V (488 nm) was used to detect apoptotic cells. To differentiate necrotic cells, PI was added (561 nm).
Lactate production
Overview
A lot of cancer cells ungergo a metabolic transition, called Warburg effect, where they do the aerobic glycolysis to obtain ATP. A subproduct they generate in this kind of energy production id L-Lactate (Vander et al, 2009). Therefore, the excretion of Lactate by cancer and healthy cells is different enough to be used as an indicator of cancer.
First, we wanted to check the how the production rates of Lactate would vary in healthy and cancer cells. To this end, we analysed the medium in which our cells of interest had been incubated for a defined time in a defined seeding density. For detection of Lactate in a sample, we used a L-lactate kit (Megazyme), in which two enzymatic reactions lead to a measurable concentration of NADH proportional to the Lactate content of the sample, which can then be measured as optical density at 340 nm (OD340). The kit was used applying the provided protocol.
Results
The first problem we came across measuring the absorbance was the phenol red of the medium, which absorbs at the same wavelength as the Lactate kit. Once the phenol red was removed, there was still another problem: a component of the medium seemed to react with our kit. After a bibliographic search, we decided that removing FBS from the medium was our best chance to get reliable calibration curves and measurements. We also incorporated a centrifugation step after the collection of the sample to remove any cell that could stay in suspension.
Lactate concentrations were measured per well, and then normalized by the number of cells present in the well (see protocol about the measurement of Lactate in mammalian cells).
Bacterial sensor
We performed a series of experiments to validate the expression of our constructs as well as the functionality of our system. Te results are presented in the following.
INP-Annexin V expression
Overview
In Microbeacon system we rely on bacteria binding to cancer cells as a co-localization signal. The solution that we propose is to use Annexin V expressed on the bacterial membrane to bind cancer cells. Annexin V binds to phosphatidylserine, which is a membrane lipid usually found in the inner part of the cell, but in cells undergoing apoptosis it flips to the outer membrane due to the activity of flipases.
Annexin V is a soluble protein with an unknown function. As we want to express Annexin V in the outer membrane we need a protein that will export it. For this purpose we chose INP ((BBa_K523013)) and changed the YFP for a human Annexin V protein optimized for Escherichia coli codon usage (Life Technologies). We used a strong RBS (BBa_B0034), a low expression promoter (BBa_J23114) and a low copy plasmid pSEVA371 to avoid an excessive stress for the cell, as it is a membrane protein.
Results
To test our construct we used:
- J23114-B0032-INP-Annexin V in pSEVA371 (low copy plasmid)expressing RFP in TOP10 cells
- J23114 expressing RFP in TOP10 cells
To test whether our experiment works, we used for colonies with the above description. Then, as a first test, we did a test with beads. We used magnetic beads, which we incubated with antibodies for 30 min and then with the bacteria with different constructs. Unfortunately, we got no results from them. We also did a Western blot using our anti-Annexin V antibody (Affimetrix BMS147) observing no band in the Annexin V weight.
We wondered what could have gone wrong, so we checked with an RBS calculator (Salis Lab) whether the expression would be high enough with our design. There, we realized that Annexin V expression was too low and probably our protein was not being expressed. Using B0032 with INP-Annexin V we were getting 410 a.u., while if we changed the RBS to B0034, we would get 12 000 a.u. Therefore, we changed the RBS of INP-Annexin V and Annexin V to B0034. As can be seen in Figure X we can see a band at the expected 35.9 kDa. This confirms the expression of Annexin V in our cells.
INP-Annexin V externalization
Overview
After proving expression of our Annexin V-INP construct it is also crucial to test ot for it's expression and functionality.
Characterization of the LldR promoter
Overview
For the sensing of Lactate inside our system we have a constitutive expression of LldR. LldR is a protein which binds to two operators, operator1 and operator2, that flank the natural LldR promoter. This interaction forms a DNA loop, which does not allow the polymerase to transcribe the respective gene. Once Lactate is added to this system, it sequester LldR which can then no longer repress the promoter, so the gene of interest (LacI in the final system) can be expressed.
An improvement to our system is the addition of LldP, a protein which is known to insert into the membrane and increase the importation rate of Lactate into the cell. either put something like "important due to modelling" or "introduced after initial findings" or rephrase this.
To find the optimal regulatory system, we designed nine versions of this discribed promoter that would be released from repression by LldR upon addition of Lactate:
Table X. Design of the promoter regulated by LldR.
| O1-promoter-O2 | promoter-O1-O2 | promoter-O1-spacer-O2 | |
|---|---|---|---|
| J23100 | p69 | p63 | - |
| J23117 | p70 | p65 | p66 |
| J23118 | p71 | p67 | p68 |
| pLldR | p62 | - | - |
For characterization of the promoter, we cloned all versions in front of a reporter-gfp. We then measured the sensitivity of our designed promoters to a range of Lactate concentrations by monitoring the intensity of gfp produced by our bacteria expressing constant LldR along with the nine reporter plasmids. Further information on the experimental setup can be found here.
We tested the following combinations of plasmids:
- a medium copy plasmid (pSEVA261) containing our designed lldR-promoters followed by a reporter gfp
- the described medium copy plasmid and a low copy plasmid (pSEVA371) containing constant lldR
- the described medium copy plasmid and a low copy plasmid containing constant lldR and constant lldP (low levels)
- the described medium copy plasmid and a low copy plasmid containing constant lldR and constant lldP (high levels)
For reasons of simplification, in the following we will referr to the genomic levels of LldR without overexpression from a plasmid as "no lldR".
Results
Initial experiments with all nine promoters showed a clear biase of functionality towards the three versions ressembling the architecture of the natural lldR-promoter (data not shown). For furtehr experiments we therefore focussed on these versions:
- p62 (natural version for reference)
- p69
- p70
- p71
We were lucky to establish a collaboration with Stockholm, where they offered to characterize the find better linkless favourable designs for us. In the following we will only focus on the four most promising versions.
We continued to characterize our reduced set of promoters by recording dose response curves amongst a broad range of Lactate concentrations. The first thing to test was wether or not our promoters actually respond to lldR as we expected them to, i.e. if gfp intensity would be properly repressed in the presence of lldR. The resulting data is shown in Figure x.
Surprisingly, what we found was not the expected release from the repression of LldR in the presence of Lactate. Indeed, if no LldR is present in the cell, according to the previous model, the levels of fluorescence should be equivalent as when there is high Lactate and LldR. We can clearly observe additional activation of GFP production! Especially for p69 and p71 the levels of normalized fluorescence upon introduction of Lactate into the system containing LldR rises way above levels observed without LldR1. This interesting finding leads us to reinterpret the present model. Instead of having only repression by LldR, it seems that LldR is required for the transcription machinery. LldR would play a dual role as an activator and as a repressor. adapt our model to accomodate this interesting, unpublished finding.
As mentioned above, we then introduced the Lactate symporter lldP into our system. Due to difficulties in cloning, expression of LldP and LldR are coupled in this case, the result of which is rather low levels of LldR in comparison to the experiment shown above in which there is no lldP (see the table below). We are comparing two different levels of lldP-LldR. To estimate which part of the observed changes results from differences in levels of LldR or lldP, respectively, we also designed a different version of LldR where the expression level is more comparable to the versions including lldP.
| Construct | Expression of LldR(A. U.) | Expression of LldP(A. U.) |
|---|---|---|
| LldR | 51100 | |
| Low LldP- lldR | 664 | 8400 |
| High LldP- lldR | 12 | 23.4 |
Comparison of low levels of LldR and LldP, to the second panel, representing high levels of LldR, can suggest either of two things:
- introduction of lldP into the system leads to leakyness due to uptake of possible traces of Lactate in the medium
- the levels of LldR are too low to properly suppress the LldR-promoter even without presence of Lactate
To discriminate these two possibilities we tested the system, as mentionned, with a higher expression of LldR and LldP (see fourth panel). The reduced level of leakyness despite higher levels of lldP suggests that the problem before was not import of Lactate from the medium (which should not be present anyways) but due to insufficient levels of LldR and therefore lack of proper repression in absence of Lactate.
Based on these results we decided to use p70 in our final system since it forms the most defined dose response curve with a Hill coefficient close to 2. Next to this, fortunately all of our promoters react with high sensitivity to Lactate levels in the range of bla to bli molar, which is the range in which we expect the Lactate output of our cancer cells to be located.
Characterization of the lacI-lldR promoter
Overview
Results
Quorum sensing modul
Overview
Results
1) For reasons of simplification we are referring to the genomic levels of lldR without overexpression from a plasmid as "no lldR".
Mammalian cells and bacteria combined
Co-culture of mammalian cells with bacteria
Overview
One issue that was concerning us was the possibility that the mammalian cells would behave in unusual ways in presence of bacteria. To exclude this possibility we decided to co-culture mammalian cells with bacteria for 3 h which is the time we expect that our test will last.
We used E. coli expresssing GFP and for the mammalian cell lines we used 3T3, HL-60 and Jurkat lines.
Results
We could observe that after the set time point the cells had the same morphology as in the control.