Team:ETH Zurich/Modeling/Reaction-diffusion
- Project
- Modeling
- Lab
- Human
Practices - Parts
- About Us
Reaction-diffusion Models
Goals
While single-cell <math alt="Square root of pi">\sqrt{\pi}</math> models are useful for correctly implementing and debugging chemical reaction models, we need to use a reaction-diffusion model to fully understand our system's functionality. Through this modeling, we aim to implement and explore
- the diffusion of AHL and lactate through the well
- the relative accessibilities of bound and unbound E. coli to lactate produced by the target cell
- whether our system self-activates in the lack of high lactate production and at what time
- the effects of E. coli population growth on the system
- comparing AHL degradation with riboregulation with respect to delaying the self-activation time
- model the binding and unbinding of E. coli to the target cell
Overview
The results from these simulations show that due to our experimental environment, the production of an AHL gradient oriented towards the outer edge of the well (i.e. higher concentration near the target cell) is not possible due to the fast diffusion of AHL. However, the greater capture of lactate by cells bound to the target relative to free-floating cells creates a concentration gradient of bound lactate and LuxR which translates to a difference in GFP signal between cells in the two locations. Combinatorially testing cases where E. coli cells bind or float around high and low lactate producers shows that our system acts as an AND gate of the two cancer markers, with E. coli bound to a high lactate producer fluorescing at orders of magnitude greater strength.
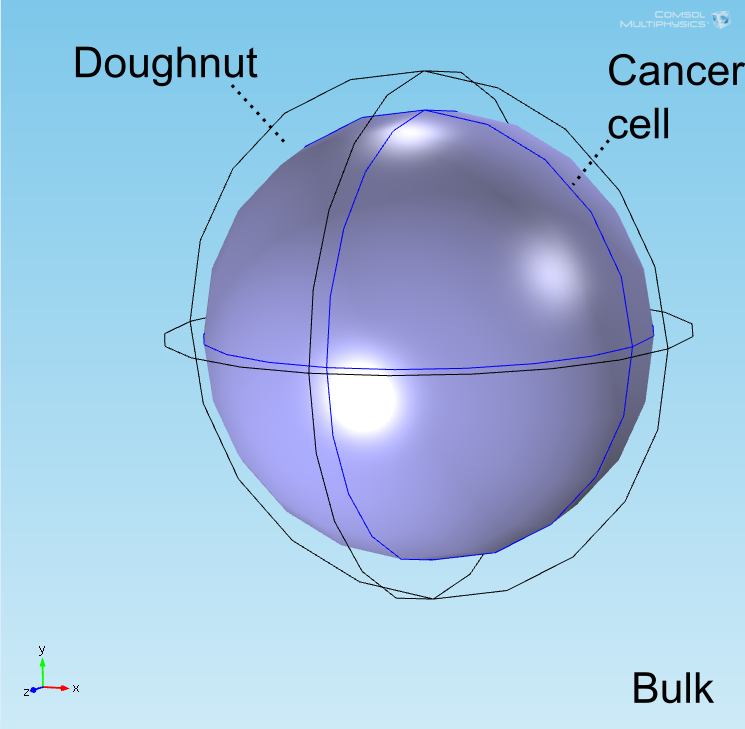
The "doughnut" model
Introduction
After implementing the single-cell models in the COMSOL multiphysics software package in an analogous geometry as a control, a more accurate geometry was constructed. Preliminary simulations were done in a geometry where a central circle representing the target cell was surrounded with smaller circles representing bound E. coli and circles uniformly distributed in the remaining space represented unbound E. coli. It was quickly realized, however, that this would not be an accurate representation of our test due to the rapid growth of the E. coli population. To address this, the discrete circles were abstracted to connected spaces representing the union of a collection of cells to allow for population growth to be simulated by modification of the ODE system over time. Owing to the shape of the layer of E. coli bound to the target cell, this geometry is referred to as the "doughnut" model. The AHL and lactate modules were implemented and tested separately before being combined into a full model.
All mathematical derivations are done in general, with values for variables being defined in the definition of our model geometry and assumptions, and in the parameters page.
Abstraction of discrete E. coli to a connected space
The well in which our reaction takes place is a box of length 200μm. In the middle of the well, we have a circle representing our target cell surrounded by a ring (called the doughnut) of width equal to the diameter of an E. coli cell representing the union of all bound E. coli cells (see Figure 1). Finally, the remaining space represents both the medium and free-floating E. coli cells, which we refer to as the bulk. All species are able to diffuse freely through this medium and rates for our reactions are adjusted based on the size of the E. coli population at a given time.
Assumptions
- Uniform distribution of cells in their respective spaces
- Cell density in the doughnut is constant and maximal
- A cell unbinding from the target is immediately replaced
- Logistic growth of E. coli in the bulk
- E. coli doubling time of 30 minutes
- One non-dividing target cell per well
- Experiment ends after twice the amount of time taken to reach the E. coli carrying capacity

Lactate Module
\begin{align} eq1 &= 1\\ eq2 &= 2\\ \end{align}AHL Module
Logistic growth of E. coli, [LuxR] as logistic function
Initial model 1: Constant LuxR, constant population size
The initial characterization of the system was done in a model in which the concentration of LuxR was constant and corresponded to one-third of its steady-state concentration. In addition, the population sizes of the E. coli in the doughnut and bulk were kept constant at 150 and 12798 cells, respectively. Under this framework, the toggling of riboregulation of the LuxI promoter was a binary parameter, while the concentration of AiiA was varied with concentrations ranging from 0 μM to 800 μM. These test are identical to those done when simulating the compartment model and were performed as a positive control of the system's implementation.
- Without AiiA, ribo
- ETH GFP ribo1 aiia0 log.sv
- ETH GFP ribo0 aiia0 log.sv
- With AiiA, ribo vs noribo (doughnut vs bulk) vs noribo (doughnut, varying Caiia)
- Error creating thumbnail: File missing
- Error creating thumbnail: File missing
- Error creating thumbnail: File missing
When a riboregulated promoter is not used for LuxI, varying the concentration of AiiA delayed the activation of the
In addition to testing the influence of riboregulation and AHL degradation, we also explored the effects that the size of the bulk population has. Starting from a size of 1000, a parameter sweep was performed in intervals of 500 cells until the E. coli carrying capacity of 12,000 cells was reached (see below). A linear decrease in the GFP concentration ratio was observed.
Initial Model 2: Constant LuxR, logistic growth of bulk E. coli
Being a more realistic model of population size, the natural extension to the above work was the introduction of a logistic growth model for the bulk E. coli population. The population was initialized to 1000 cells and allowed to grow until the well's carrying capacity. The new growth model did not significantly change the final ratio of GFP concentrations, but instead led to a slower activation of the system in the first 300 minutes.
- Ribo, constant pop vs logistic growth
- Error creating thumbnail: File missing
- Error creating thumbnail: File missing
Full Model
Conclusions
By modeling the diffusion of lactate through our well, we have demonstrated that the E. coli bound to their target cell, through active transport and capture by binding to LldR, reduce the accessible lactate concentration of the unbound cells, producing a concentration gradient of its output signal LuxR.
Simulations of the AHL module demonstrated that the leakiness of the LuxR promoter leads to the self-activation of this module within our experiment's timeframe. The degradation of AHL via the action of AiiA did not lead to a drastic change in this behavior, while the introduction of riboregulation to the LuxR promoter tied to LuxI suppressed self-activation sufficiently.
Model Details
Logistic growth of E. coli
Let us define \(n_\text{bulk}:[0,t_\text{sim}]\longrightarrow \mathbb N\) as the function representing the size of the E. coli population in the bulk at time \(t\), with an initial population size of \(n_0\), a carrying capacity of \(K\), and a growth rate of \(R\). The closed-form equation of this function is then $$n_\text{bulk}(t) = \frac{n_0 K e^{Rt}}{K + n_0(e^{Rt}-1)}$$
If we denote the doubling time of the population by \(t_2\), we can solve for the growth rate \(R\) and define a new constant \(g\) s.t. $$R = \frac{\log2 + \log(K+n_0) - \log(K-2n_0)}{t_2} =: \frac{\log g}{t_2}$$ When we plug this value back into the original equation, it simplifies to $$n_\text{bulk}(t) = \frac{n_0 K g^{\frac{t}{t_2}}}{K + n_0(g^{\frac{t}{t_2}}-1)}$$ Finally, if we define the time we end our experiment, denoted \(t_\text{sim}\), as the time when the population reaches size \((1-\varepsilon)K\) for some small \(\varepsilon>0\), then we can solve for \(t_\text{sim}\) $$t_\text{sim} = \frac{1}{R}\log\frac{(K-n_0)(1-\varepsilon)}{n_0\varepsilon}$$
Carrying capacity
Some stuff here about the carrying capacity.

Diffusion and transport of chemical species
Under alkaline conditions, E. coli actively import lactate via a proton-motive symporter lldP.[] Thus, a cross-membrane transport reaction had to be implemented. Since diffusion across barriers is implemented with equal diffusion coefficients for both directions in COMSOL, the transport of lactate across the doughnut membrane was implemented by modeling two different states of unbound lactate. If we let our reference space be the interior of the doughnut, we can define two pseudo-species \(Lact_\text{in}\) and \(Lact_\text{out}\), denoting intracellular and extracellular lactate, respectively. \(Lact_\text{out}\) is produced by the target cell and can diffuse freely though the medium and all membranes, while \(Lact_\text{in}\) is in equilibrium with \(Lact_\text{out}\) in the doughnut with rate constants set to maintain a 20-fold difference of lactate concentration between interior and exterior. $$ Lact_\text{out} \mathop{\mathop{\xrightarrow{\hspace{4em}}}^{\xleftarrow{\hspace{4em}}}}_{k_\text{in}}^{k_{\mathrm{ext}}} Lact_\text{in} \qquad \frac{k_\text{in}}{k_\text{out}}\approx 20 $$ and cannot diffuse out to maintain its high concentration in the doughnut. Only the \(Lact_\text{in}\) state can react with the other chemical species in the doughnut and only \(Lact_\text{out}\) can react with other chemical species in the bulk.
Since AHL can freely diffuse across membranes, unbound AHL is only modeled as one state that can diffuse across all barriers. The effective diffusion coefficients of AHL and Lactate through the doughnut membrane \(D_e\) were approximated relative to their coefficients in water \(D_{aq}\) by the relation $$\frac{D_e}{D_{aq}}\approx 0.25$$ using the approximation proposed in a 2003 review by Stewart.
Correcting concentrations
Let \(\mathbf X:[0,t_\text{sim}]\longrightarrow \mathbb R^n\) be a function representing the molar concentrations of our chemical species in a real cell over the simulation period with units \(\frac{\text{mol}}{L}\) for each component of \(\mathbf X\). For a detailed description of the system, its assumptions and its derivation, please see the pages on the lactate module, AHL module and the combined model.
Since the areas of the doughnut and bulk are not equal to the areas of the spaces they are representing, a new ODE system equivalent to our model, but occurring in the model spaces, needs to be calculated. We define the function \(f\) such that our ODE system can be represented in the form \(\left.\frac{d\mathbf X}{dt}\right|_{(\mathbf X(t),t)} = f(\mathbf X(t),t)\). We can then define a new function \(\mathbf Y(t) := \nu(t)\mathbf X(t)\) representing the concentrations of the species within our model spaces, where $$\nu_\text{bulk}(t) := \frac{n(t)A_\textit{E. coli}}{A_\text{bulk}}\qquad \nu_\text{doughnut}(t) := \frac{n_\text{doughnut}A_\textit{E. coli}}{A_\text{doughnut}}$$ are concentration correction factors corresponding to our abstract spaces. Solving for \(\frac{d\mathbf Y}{dt}\) yields for each space yields \begin{align*} \left.\frac{d\mathbf Y_\text{bulk}}{dt}\right|_{(\mathbf Y(t),t)} &= \nu_\text{bulk}(t)\left(f\left(\frac{\mathbf Y(t)}{\nu_\text{bulk}(t)},t\right) - R\left(\frac{n(t)}{K}-1\right)\mathbf Y(t)\right)\\ \left.\frac{d\mathbf Y_\text{doughnut}}{dt}\right|_{(\mathbf Y(t),t)} &= \frac{n_\text{bulk}A_\textit{E. coli}}{A_\text{doughnut}}\left(f\left(\frac{A_\text{doughnut}}{n_\text{doughnut}A_\textit{E. coli}}\mathbf Y(t),t\right)\right) \end{align*} Since \(AHL\) and \(Lact_\text{out}\) are the only species that are allowed to diffuse freely between the cells, we can obtain an equivalent system operating in the model spaces by multiplying the concentrations of AHL and \(Lact_\text{out}\) by \(\nu\) and dividing their derivatives by \(\nu\) to save on computing time.
Binding and unbinding of cells
None of the other chemical species are able to significantly diffuse across cell membranes in vivo. However, in order to model the unbinding and binding of cells to the target cell, an approach of implementing separate intracellular and extracellular pseudo-species was applied to the remaining chemical species. $$ X^i_\text{out} \mathop{\mathop{\xrightarrow{\hspace{4em}}}^{\xleftarrow{\hspace{4em}}}}_{k_\text{in}}^{k_{\mathrm{off}}} X^i_\text{in} $$ Unlike the model for lactate, the reaction rates for this equilibrium are governed not only by the concentrations of the relevant chemical species, but also by the number of cells that are part of the doughnut. So, a chemical analog of cell count has to be modelled. For this section, we are no longer assuming that the density of the doughnut is constant and maximal.
Binding of E. coli cells is through the binding of surface-exposed annexin V (A5) on their membrane to phosphatidylserine (PS) on the target cell surface to form the complex (APS). $$ A5 + PS \mathop{\mathop{\xrightarrow{\hspace{4em}}}^{\xleftarrow{\hspace{4em}}}}_{k_\text{on}}^{k_{\mathrm{off}}} APS $$ By our assumption that the target cell does not divide, the total amount of PS in the system is constant (\([PS]_t = [PS] + [APS]\)). If we let \([A5]_s\) denote the annexin V concentration per cell, the system can then be modeled by the ODE $$\frac{d[APS]}{dt} = k_\text{on}[A5]_s n_\text{bulk}(t)([PS]_t - [APS]) - k_\text{off}[APS]$$ If we assume that each E. coli cell has an equal amount of annexin V on its surface, then we can define the cell count as $$C(t) = \frac{[APS]}{[PS]_t}n_\text{around}$$ and an ODE system for the cell count $$\frac{dC}{dt} = k_\text{on}[A5]_s n_\text{bulk}(t)(n_\text{around} - C(t)) - k_\text{off}C(t)$$ Finally, we can use this to define an ODE system for the conversion of the intracellular and extracellular states of our chemical species $$\frac{dX^i_\text{out}}{dt} = k_\text{off}X^i_\text{in}C(t) - \alpha(t) k_\text{on}[A5]_sn_\text{bulk}(t)X^i_\text{out}(n_\text{around}-C(t))$$ where \(\alpha(t)\) is a rate correction term that has not yet been defined (will it ever be? who knows?).
Testing our system's logic
Ideally, our system should produce much stronger fluorescence only in the presence of both input signals (higher lactate production and co-localization signals). To test whether our system acts as an AND gate on these signals, we combinatorially checked all four cases of environments with high vs. low lactate production and E. coli co-localization vs. uniform distribution, along with a fifth well with no target cell present. Although the positive feedback loop of the AHL module will eventually activate in all cases due to the leakiness of pLuxR, it was expected that a large time delay would exist between the high lactate and colocalization test and all others, or that a greater feedback would be produced in that environment.
Results
Effects of colocalization
Although our original plan was to strengthen quorum sensing between the doughnut cells through AHL degradation in the bulk, it became apparent that the high diffusion coefficient of AHL and the small size of our well prevented this from occurring. Within a one minute period, any produced AHL quickly diffused to produce a uniform distribution. The introduction of increased AHL degradation in the bulk led to an increased rate of AHL diffusion into the bulk and did not produce a gradient as desired.
Fortunately, the co-localization of cells in the doughnut produced a substantial lactate and LuxR gradient due to lactate capture. Since pLuxR is tied to both LuxI and GFP, this concentration gradient produces a stronger positive feedback response in these two proteins, thus producing the desired GFP concentration gradient.
Our system as an AND gate