Team:Czech Republic/Project
Project
Contents
Abstract
The IOD band is a general diagnostic test enabling early detection and mapping of tumor mobility. Over a billion unique tests are made accessible to field experts outside of synthetic biology with a unique clone-free assembly feature. Tumor mobility is incredibly difficult to diagnose due to the rarity of circulating tumor cells (CTCs) and the complexity of surface marker combinations. The IOD band strives to make it easy. The central players are processing units called Input Output Diploids or IODs. IODs use antigen recognition and intercellular communication to create a logical network by which even single cells carrying the desired marker profile can be identified in a background of millions. Affirmative CTC localisation triggers a global response manifested by IOD initiated clumping at levels visible to the naked eye. As such, IOD bands do in a test tube what normally requires days to do in the lab.
Motivation
Tumor mobility is likely the most significant prognostic factor for all types of cancer. Contained primary tumors often present no symptoms and if discovered early can be safely removed without needing subsequent chemotherapy. If left untreated, however, primary tumors spread through the lymphatic or blood circulatory systems to other parts of the body. Given enough time, the cancer cells transition and take on the forms of cells from other organs. At this stage, the cells invade compatible organs and secondary tumors called Mets develop. Early mets are less diverse and still present hope for treatment. Later mets, however, are too diverse and are usually associated with terminal diagnosis.
The difference between early stage and late stage diagnosis can be staggering. The table below lists the survival rate differential between early and late diagnosis for common cancer types.
| Stage | Kidney | Breast | Lung | Colorectal | Skin | Prostate |
|---|---|---|---|---|---|---|
| Stage I | 81% | 100% | 45% | 92% | 86% | 100% |
| Stage IV | 8% | 22% | 1% | 11% | 15% | 28% |
The IOD band concept
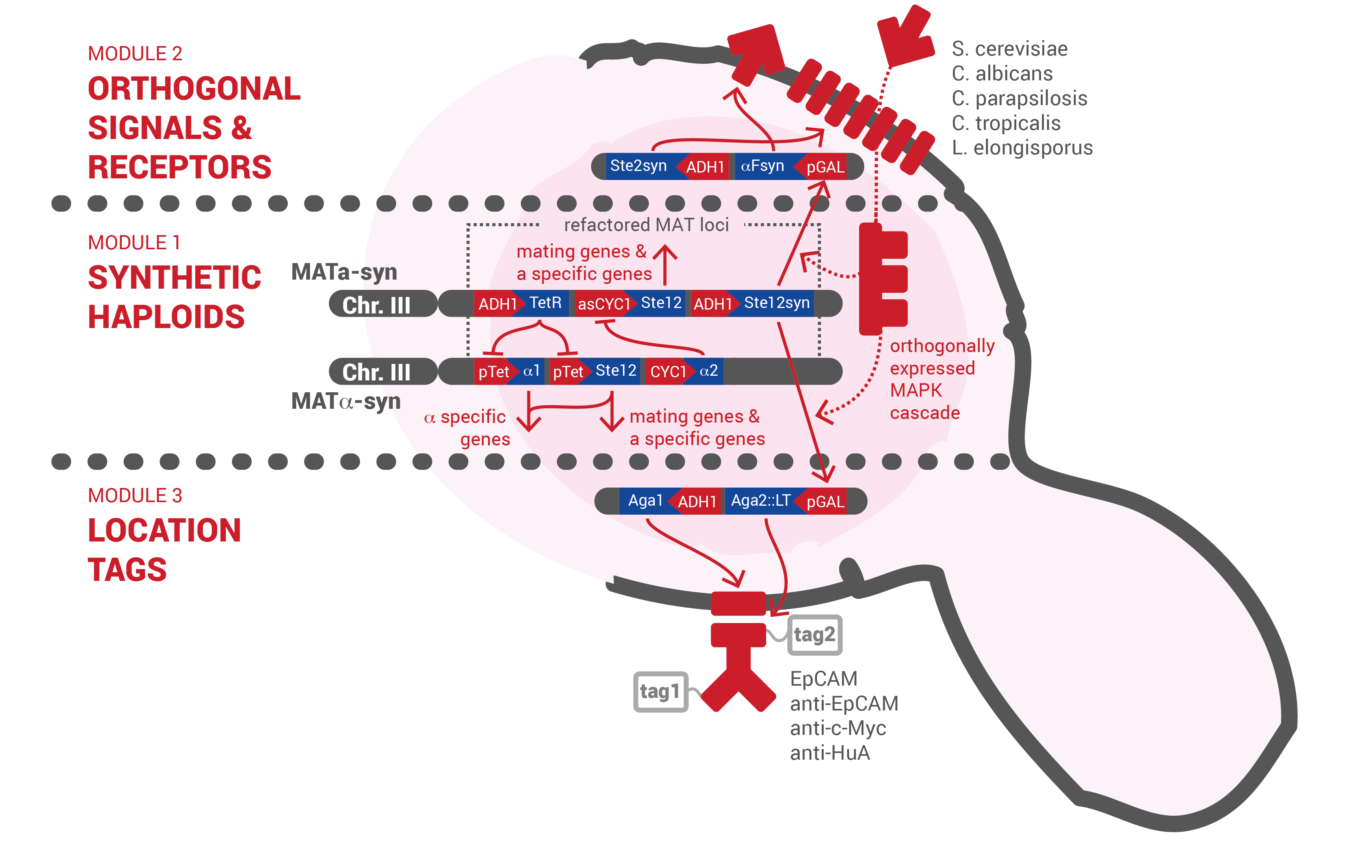
At Team Czech Republic, we envisioned a tumor marker body atlas. A large number of tumor cell lines as well as healthy tissue of common secondary sites have well documented surface marker profiles. A dataset of over 700 surface markers comprising common receptor, transmembrane protein, and channel families was processed to identify usable members with confirmed antibodies. Each cell line was then linked to a unique combination of up to three markers identified by a simple 6 or 9 digit barcode. We then envisioned The IOD Band, a marker profile probe that locates cells with target marker combinations in samples of peripheral blood.
The fundamental units of the IOD band are intelligent input/output diploid cells called IODs. An IOD is fundamentally simple comprising two modular inputs and a single modular output. The inputs are locational tags displayed on the surface and pheromones detected by orthogonally expressed receptor and signalling pathway. The inputs combine to yield locally limited cell-cell communication. Pheromones are also the IOD outputs. A rewiring of the signalling pathway transcription factor connects the inputs with the outputs. Hence a single IOD simply detects a pheromone signal and generates an orthogonal signal in response.
Design
The IOD band is a project built on system level integration. At the outset it seemed like an impossible goal for a single iGEM project especially as there are no precedents for many of the system parts. So the idea was stripped to its brass tacks to uncover three fundamental modules that prove the diagnostic and engineering principles in a tractable manner.
Module 1 builds synthetic haploid strains with refactored mating loci that are conjugated to make a functional IOD. These strains have the wild type mating phenotype and differentially express a reprogrammed signalling pathway in their diploid state proving the feasibility of the clone-free assembly concept.
Module 2 builds a set of orthogonal pheromones and receptors. These pheromone-receptor pairs enable the realization of multichannel signal transmission required to implement logic operations necessary for reliable diagnosis.
Module 3 builds a set of locational tags that recognise common tumor surface markers and agglutinate cell populations. Location tags displayed in the correct confirmation strengthen cell-cell interactions to enable localisation of signal transmission.
In addition, modeling and microlfuidics were extensively used to support the experimental work. Modeling proved robustness of simple signal transmission networks in a simulation setting with naturally occurring signalling noise. Self replicated microfluidics chips provided the prices environment necessary to quantify the individual cell-cell interactions.
Module 1 - Synthetic haploids
Abstract
The yeast pheromone pathway is a naturally present mechanism of signal transduction in yeast cells. In order for IODs to use mating pheromones as signalling molecules, each IOD has to have a mechanism to process input signals. We would like to use the modified pheromone pathway in our IODs, however in diploid cell all components of the pathway are naturally switched-off, therefore we designed synthetic haploid strains of both mating types that preserve the ability to process an extracellular signal via pheromone response pathway even after mating - in diploid state. As a result, IODs use the robust signaling pathway for signal processing.
Key Achievements
- Constructed a set of reporter promoters for yeast cells.
- Characterized reporter promoters in all mating types
- Designed and materialized synthetic MATa and MATx strains
- Built a synthetic diploid strain with a functional yeast pheromone pathway
- Demonstrated the correct functionality of yeast pheromone pathway in synthetic diploids
Module 2 - Orthogonal signals and receptors
Abstract
In order to achieve more complex behaviour of our IODs, each IOD needs to be able to communicate with others. Since the pheromone response pathway was used for signal transduction, we prepared several synthetic signals and receptors that couple to this pathway and allow our IODs to communicate without a cross-talk.
Key Achievements
- Constructed a set of yeast plasmids with different mating pheromones and their receptors.
- Verified the correct coupling of the receptors to the yeast pheromone mating pathway.
- Verified the correct expression and secretion of the different pheromones.
Module 3 - Location tags
Abstract
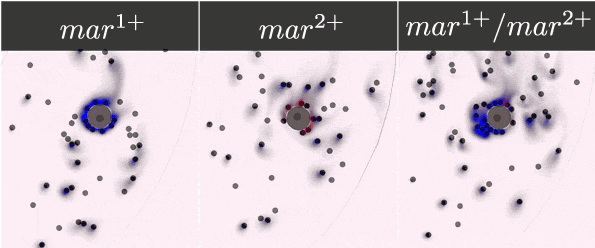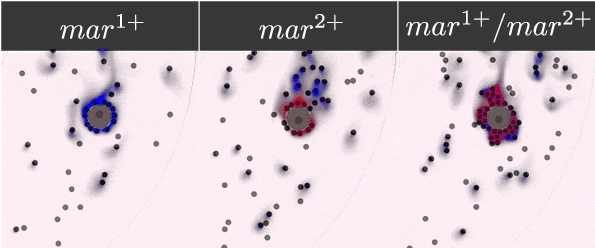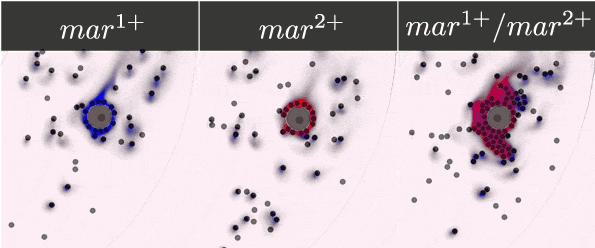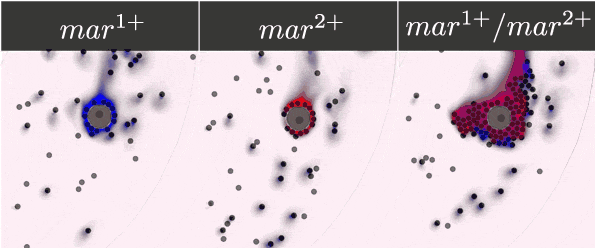
To guarantee the ability of our system to recognise, bind, and respond to the presence of a tumour cell, we selected five antibody fragments to display on our yeast cells using the yeast display technology and monitor the cells' binding. Their expression was detected and monitored on a microfluidic chip with the use of antibody staining and fluorescence microscopy.
Key Achievements
- Expressed streptavidin, EpCAM, Anti-EpCAM, c-myc and HuA receptors on the surface of yeasts.
- Monitored the dynamic binding of our receptors and corresponding markers.
- Demonstrated the ability of our receptors to bind chosen markers.
Module 4 - Modelling
Abstract
IOD systems function at both the intracellular and intercellular level. Intracellular biochemical reactions determine signalling activities and gene expression. Extracellular reactions include signalling molecule diffusion and cell movement with cell-cell interactions. At each level models of various complexity are available. We selected minimalistic models that capture only the key design elements and integrated these models in a single simulator CeCe. Subsequently simulation was used to study the robustness and efficiency of different signal transmission patters in identifying cells with specific marker profiles while minimising false positives.
Key Achievements
- Developed a simulation environment CeCe to capture the complexity of cell-cell signal transmission.
- Designed an IOD chemical reaction network model.
- Developed a schematic architecture for conceptual modelling of signal transmission networks.
- Designed a two IOD signal transmission network suitable for the IOD band.
- Illustrated the robustness and efficiency of the IOD band design in CeCe simulations.
Module 5 - Microfluidics
Abstract
Microfluidic devices were designed and fabricated to characterise the signal transduction in the developed IOD band system. Signal transduction between transmitting and receiving cells was characterised using spatially separated cell cultures inside single microfluidic channel in conjunction with live fluorescence microscopy. Plasmids coding a synthetic reporter protein were transformed to signal receiving cells to show activation of its MAPK cascade. The on-chip characterisation was used to evaluate the signal transduction distance limit, its dynamic behavior, and reliability.
Key Achievements
- Set of microfluidic devices fabricated by PDMS soft-lithography.
- Characterisation of signal transduction distance limit between wildtype MATa and MATx Saccharomyces cerevisiae cells.
- Dynamic characterisation of signal transduction between synthetic MATa and MATx Saccharomyces cerevisiae cells.