Team:UChicago/Composite Part
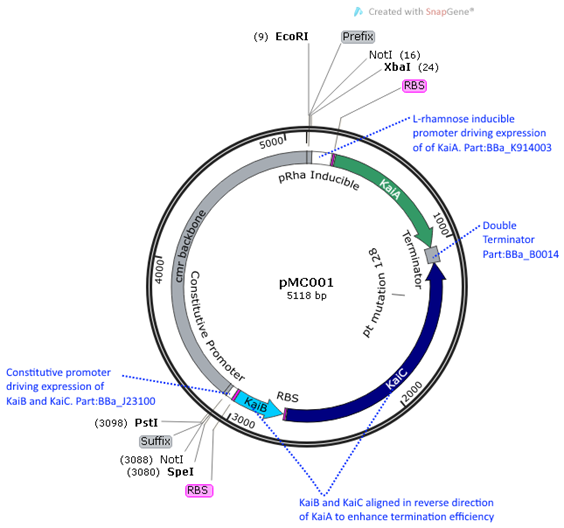
 pMC001 (BBa_K1745001) is the main Kai ABC oscillator biobrick
we submitted into the iGEM registry. It contains the
three Kai proteins -KaiA, KaiB,
and KaiC under the expression of two different
promoters. KaiB and KaiC
are under a constitutive promoter (Part J23100) and KaiA
is under a L-rhamnose
inducible promoter (Part K914003). The inducible promoter was chosen for KaiA in order to investigate the effect of altering the
stoichiometric ratios between KaiA and KaiC. This ratio was found to be particularly significant
in in vitro reactions, where the ratio of KaiA to KaiC was 1:3 (Nakajima, Science 2015).
pMC001 (BBa_K1745001) is the main Kai ABC oscillator biobrick
we submitted into the iGEM registry. It contains the
three Kai proteins -KaiA, KaiB,
and KaiC under the expression of two different
promoters. KaiB and KaiC
are under a constitutive promoter (Part J23100) and KaiA
is under a L-rhamnose
inducible promoter (Part K914003). The inducible promoter was chosen for KaiA in order to investigate the effect of altering the
stoichiometric ratios between KaiA and KaiC. This ratio was found to be particularly significant
in in vitro reactions, where the ratio of KaiA to KaiC was 1:3 (Nakajima, Science 2015).

pMC002 is one of the constructs for the
read-out system. It contains the circadian promoter kaibc
which is driven by the master transcription factor RpaA.
RpaB is included as some studies have reported both RpaA and RpaB necessary for kaibc expression (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3406716/ Hanoaka,
2012). This was also to reconstitute the system used by the Silver lab. SasA, the RpaA activator is
included as it directly binds to KaiC and
phosphorylates RpaA, linking the KaiABC
oscillator to an expression based read-out system. GFP is placed downstream the
circadian promoter, acting as a reporter for kaibc
expression. The GFP is LVA-Stop tagged in order to promote degradation, which
is useful to quantify increases and decreases of GFP which we expect to see
throughout the KaiC oscillations.

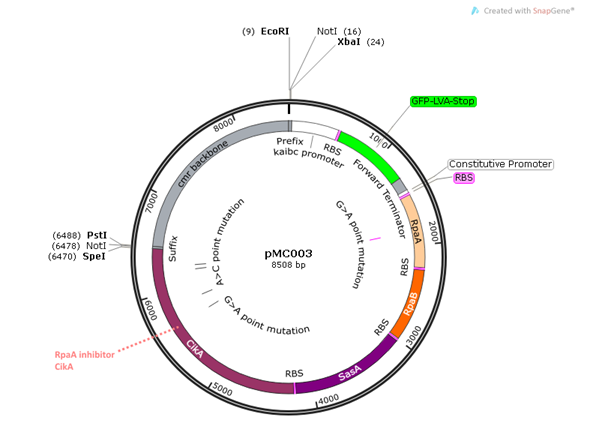
pMC003 is another read-out plasmid that will
be assayed independently of pMC002. This construct exhibits the same features
as pMC002 with the addition of CikA, a negative
regulator of RpaA which interacts directly with KaiB and dephosphorylates RpaA.
We wanted to directly examine the effects of the read-out system with and
without the CikA negative regulator to test the
hypothesis that the inclusion of CikA leads to more
robust regulation of RpaA.
Both pMC002 and pMC003 will be transformed in vitro with pMC004-7,
representing the different phosphomimetics of KaiC.
 pMC004-7 are supplemental plasmids to pMC002
and 3. They contain KaiB and KaiC
phosphomimetics. The phosphomimetics
represent different phosphorylation states of KaiC
and emulate the oscillations of KaiC. KaiB is included as it interacts directly with CikA, the RpaA negative
regulator. The phosphomimetics were generated by
altering the amino acids at position 431 and 432: AA (unphosphorylated)
(pMC004), AE (pMC005), EA (partially phosphorylated) (pMC006), EE (fully
phosphorylated) (pMC007).
pMC004-7 are supplemental plasmids to pMC002
and 3. They contain KaiB and KaiC
phosphomimetics. The phosphomimetics
represent different phosphorylation states of KaiC
and emulate the oscillations of KaiC. KaiB is included as it interacts directly with CikA, the RpaA negative
regulator. The phosphomimetics were generated by
altering the amino acids at position 431 and 432: AA (unphosphorylated)
(pMC004), AE (pMC005), EA (partially phosphorylated) (pMC006), EE (fully
phosphorylated) (pMC007).
***Note: Several parts that have no BBa plasmid
were not submitted. We have several of these plasmids not in the registry on
hand, but we forgot to put them onto a chloramphenicol backbone for submission.
Like our team Facebook page, Genehackers@UChicago!
Questions? Comments? Send us an email!
