Difference between revisions of "Team:ETH Zurich/Lab"
| (15 intermediate revisions by 7 users not shown) | |||
| Line 18: | Line 18: | ||
<h2>Overview</h2> | <h2>Overview</h2> | ||
| − | <p>In the wet lab we worked with < | + | <p>In the wet lab we worked with <a href="https://2015.igem.org/Team:ETH_Zurich/Results#_Experiments_involving_mammalian_cells_">mammalian cell culture</a>, did a lot of experiments and cloning with <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Bacterial_sensor"><i>E. coli</i></a>, developed and tested a <a href="https://2015.igem.org/Team:ETH_Zurich/Chip">microfluidics chip</a> and, last but not least, we <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Mammalian_cells_and_bacteria_combined">combined all these three parts</a> to test our system. We divide the different experiments into four sections as shown below.</p> |
| + | </html> | ||
| + | [[File:LabOverview.png|thumb|center|550px|<b>Figure 1.</b> [[Team:ETH_Zurich/Results|Overview]] of the different kinds of experiments we did.]] | ||
| + | <html> | ||
| + | |||
| + | <p>We invested a lot of time and effort into the generation of reliable data sets and never stopped looking for more experiments that would make it possible for us to achieve the most fruitful interaction with the <a href="https://2015.igem.org/Team:ETH_Zurich/Modeling">modeling</a> team, which in turn helped us a lot in setting a focus on critical points.</p> | ||
| + | |||
| + | |||
| + | </html> | ||
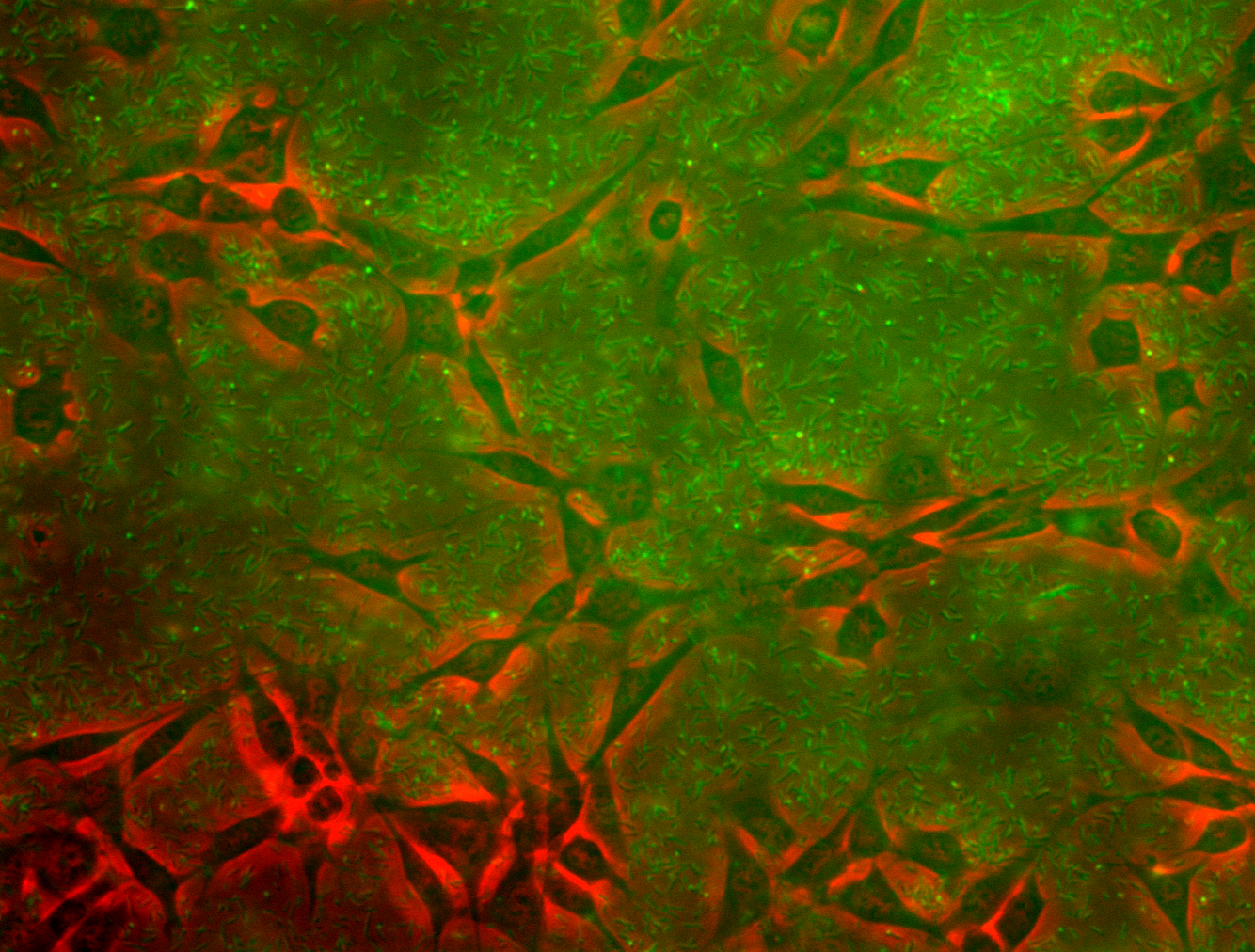
| + | [[File:ETH15_coculture_overview.png|thumb|right|[[Team:ETH_Zurich/Results#Mammalian_cells_and_bacteria_combined|Coculture]] of mammalian cells with <i>E. coli</i>.]] | ||
| + | <html> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<p>Please take a look at our experimental <a href="https://2015.igem.org/Team:ETH_Zurich/Results">Results</a> to see how we achieved</p> | <p>Please take a look at our experimental <a href="https://2015.igem.org/Team:ETH_Zurich/Results">Results</a> to see how we achieved</p> | ||
<ul> | <ul> | ||
| − | <li>the characterization of 15 newly designed <a href="https://2015.igem.org/Team:ETH_Zurich/Results# | + | <li>the characterization of 15 newly designed <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Characterization_of_synthetic_promoter_library">promoters controlled by the regulator LldR and its inhibitor L-lactate</a></li> |
<li>the characterization of 2 newly designed <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Characterization_of_the_lacI-lldR_promoter">hybrid promoters</a>, combining repression by LldR and LacI</li> | <li>the characterization of 2 newly designed <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Characterization_of_the_lacI-lldR_promoter">hybrid promoters</a>, combining repression by LldR and LacI</li> | ||
| − | <li>the closer elucidation of the <b>poorly characterized</b> mechanism of action behind the <a href="https://2015.igem.org/Team:ETH_Zurich/ | + | <li>the closer elucidation of the <b>poorly characterized</b> mechanism of action behind the <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Characterization_of_the_natural_LldR_promoter">LldR-mediated repression</a> of the LldR-operator-promoter</li> |
<li>the investigation of <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Mammalian_cells_and_bacteria_combined">binding of bacteria to apoptotic mammalian cells</a></li> | <li>the investigation of <a href="https://2015.igem.org/Team:ETH_Zurich/Results#Mammalian_cells_and_bacteria_combined">binding of bacteria to apoptotic mammalian cells</a></li> | ||
| − | |||
</ul> | </ul> | ||
<p></p> | <p></p> | ||
| − | <p>Moreover, we are | + | <p>Moreover, we are designed a testing system of our final test setup in a <a href="https://2015.igem.org/Team:ETH_Zurich/Chip">Microfluidic Chip</a>. More information about the performed experiments can be found on our <a href="https://2015.igem.org/Team:ETH_Zurich/Experiments">experiments page</a>. In addition, an overview over the bacterial strains and mammalian cell lines we used for our experiments, along with a list of materials and equipment can be found in our <a href="https://2015.igem.org/Team:ETH_Zurich/Materials">materials page</a>.</p> |
| − | <p>We are one of 71 teams that participated at this years <a href="https://2015.igem.org/Team:ETH_Zurich/Interlab_study">Interlab Study</a>, in which we | + | <p>We are one of 71 teams that participated at this years <a href="https://2015.igem.org/Team:ETH_Zurich/Interlab_study">Interlab Study</a>, in which we measured the fluorescence of the three test constructs using FACS flow cytometry, as well as our microtiter plate reader.</p> |
Latest revision as of 03:24, 19 September 2015
- Project
- Modeling
- Lab
- Human
Practices - Parts
- About Us
Lab
Overview
In the wet lab we worked with mammalian cell culture, did a lot of experiments and cloning with E. coli, developed and tested a microfluidics chip and, last but not least, we combined all these three parts to test our system. We divide the different experiments into four sections as shown below.
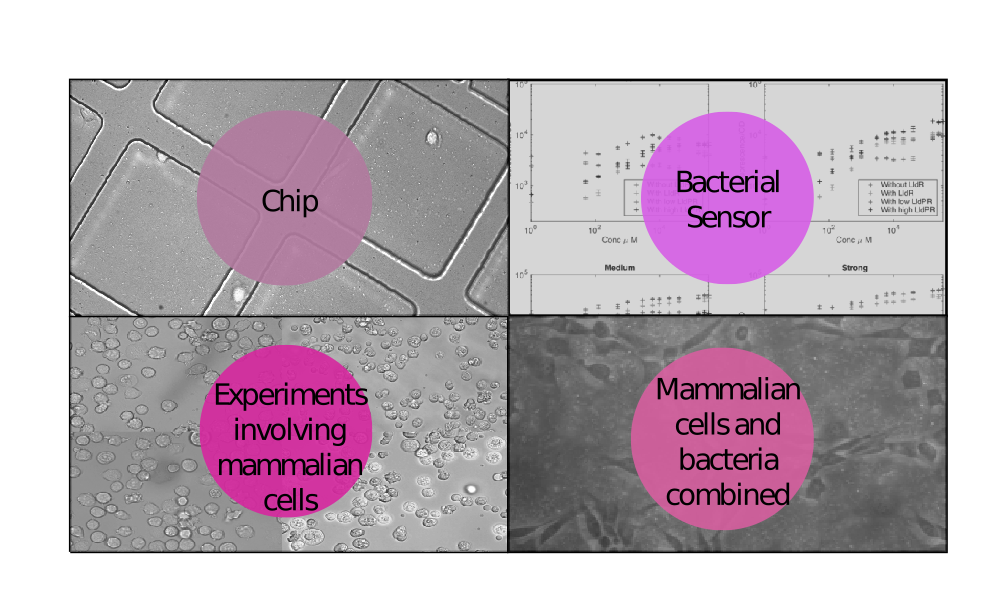
We invested a lot of time and effort into the generation of reliable data sets and never stopped looking for more experiments that would make it possible for us to achieve the most fruitful interaction with the modeling team, which in turn helped us a lot in setting a focus on critical points.
Please take a look at our experimental Results to see how we achieved
- the characterization of 15 newly designed promoters controlled by the regulator LldR and its inhibitor L-lactate
- the characterization of 2 newly designed hybrid promoters, combining repression by LldR and LacI
- the closer elucidation of the poorly characterized mechanism of action behind the LldR-mediated repression of the LldR-operator-promoter
- the investigation of binding of bacteria to apoptotic mammalian cells
Moreover, we are designed a testing system of our final test setup in a Microfluidic Chip. More information about the performed experiments can be found on our experiments page. In addition, an overview over the bacterial strains and mammalian cell lines we used for our experiments, along with a list of materials and equipment can be found in our materials page.
We are one of 71 teams that participated at this years Interlab Study, in which we measured the fluorescence of the three test constructs using FACS flow cytometry, as well as our microtiter plate reader.





















