Team:Nagahama/Medal Parts
Contents
- 1 BioBrick Parts to achieve each medal requirement
- 1.1 Bronze
- 1.2 Silver
- 1.2.1 Terpene precursor mass-production device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653024 BBa_K1653024])
- 1.2.2 Farnesol (FOH) production device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653025 BBa_K1653025])
- 1.2.3 Geraniol (GOH) production device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653027 BBa_K1653027])
- 1.3 Gold
BioBrick Parts to achieve each medal requirement
Bronze
We created 30 new BioBricks[1] for bronze medal criterion. And have documented and submitted them to igem Registry.
We introduce ispA ([http://parts.igem.org/Part:BBa_K1653003 BBa_K1653003]) of them. This biobrick is Key BioBrics in "Flavorator" project.
ispA encodes Farnesyl diphosphate synthase. Farnesyl diphosphate synthase can utilize both dimethylallyl and geranyl diphosphates as substrates, generating geranyl and farnesyl diphosphate, respectively. Therefore the enzyme can catalyze two sequential reactions in the polyisoprenoid biosynthetic pathway.
Silver
We created 3 new BioBrick devices for silver medal criterion.
Terpene precursor mass-production device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653024 BBa_K1653024])
To realize "Flovorator" we need to make E. coli produce a large amount of terpene precursor. Surprisingly, We have succeeded in its mass-production by terpene precusor mass-production device. Our new biobrick device worked as expected. Because we validate this fact by ubiquinone thin-layer chromatography (TLC) analysis experimentally. We have documented and submitted this new Biobrick device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653024 BBa_K1653024]) to iGEM Registry .
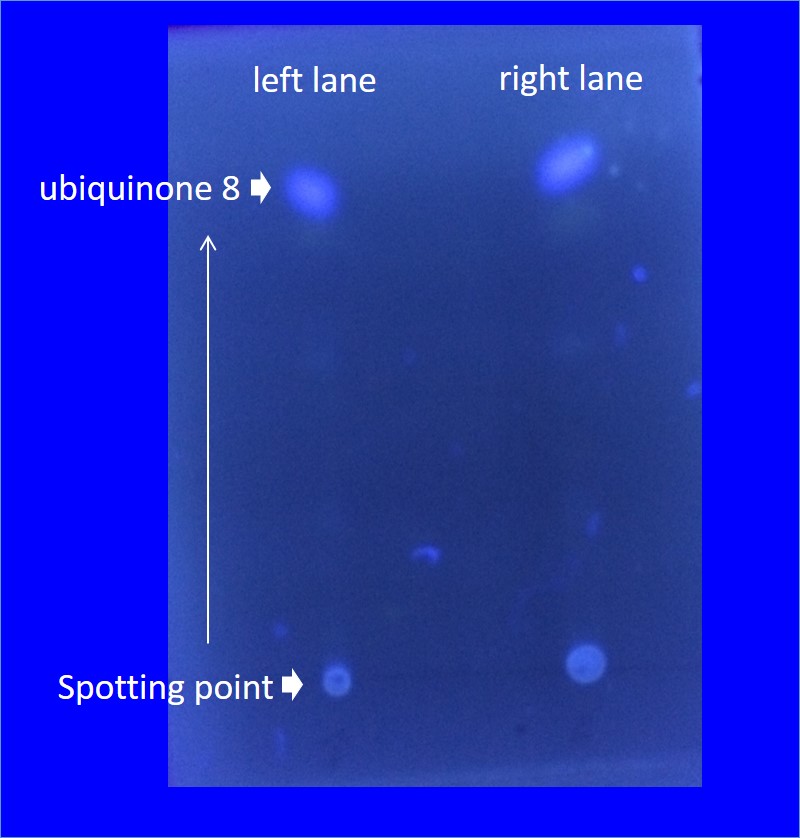
We want to make the E.coli produces farnesol and geraniol which are one of the terpenoids. To produce great quantity of terpenoids they need many terpene precursor. E. coli produces a small amount of the terpene precursor in MEP pathway. In MEP pathway, there are four enzymes (ispD, ispf, idi, dxs) which are speed limiting enzyme for terpenoids precursors produce in E. coli. In order to create a high-yield strains producing IPP and DMAPP, we exogenously engineer to superimpose these genes into E. coli to create strains overproducing IPP and DMAPP in a MEP pathway.To confirm increased production of terpene precursors by Terpene precursor mass-production device. we put attention on ubiquinone. Ubiquinone 8 is made from Farnesyl diphosphate (FPP) which is one of the terpene precursors . quinone is one of the electron carrier present in the cell membrane of prokaryotes. And also they glow when exposed to UV rays.In the measurement of production of quinone it was measured by thin-layer chromatography. (TLC silica gel)
Result:
The figure on the left is analysis of ubiquinone 8 by thin-layer chromatography. Right lane is JM109 /Terpene precursor mass-production device with IPTG Left lane is JM109/Terpene precursor mass-production device IPTG minus . Both it was 2μl spot.
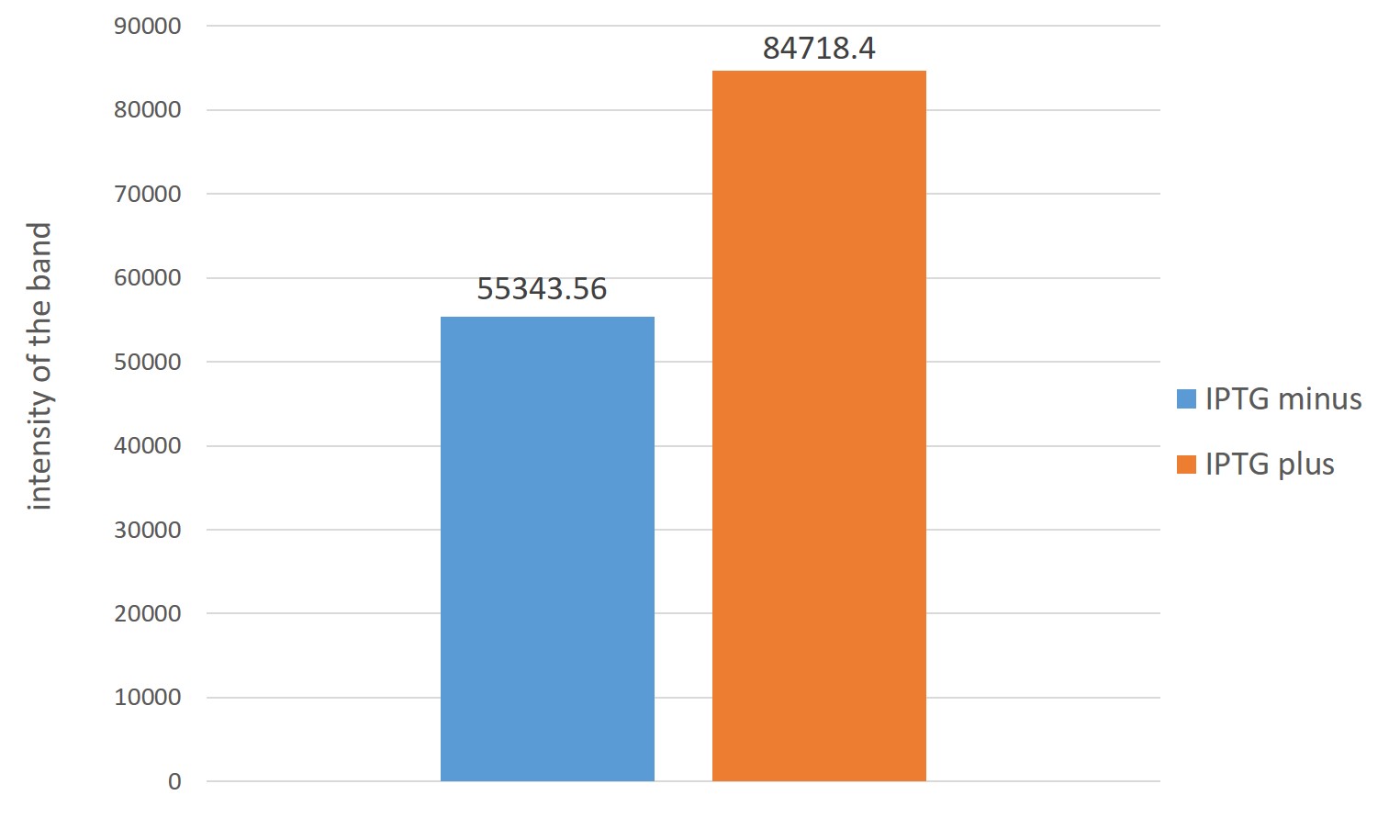
The right of the figure,Estimation of ubiquinone-8 content instead Each intensity of spots indicating the content of ubiquinone-8
From two figures, those which are overexpressed in reintroduced to E. coli four genes, it is better to have overexpressed were many production of ubiquinone 8 as compared with those that do not overexpress
Discussion:
From this result, the amount of ubiquinone 8 of the final material by the increased amount of terpene precursors is increased by re-introducing the four genes are over-expressed in E. coli.
Therefore, it considered could strengthen the MEP pathway.
Farnesol (FOH) production device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653025 BBa_K1653025])
To realize "Flovorator" we need to make E. coli produce Farnesol. Surprisingly, We have succeeded in farnesol production by Farnesol production device.
Our new biobrick device "Farnesol production device" worked as expected. Because we validate this fact by GC-MS analysis experimentally. We have documented and submitted this new Biobrick device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653025 BBa_K1653025]) to iGEM Registry .

FOH is probably generated through FPP hydrolysis by endogenous phosphatases, which are induced by an increased intracellular FPP level. Analogously, we hypothesized that E. coli could produce FOH under cellular conditions of an increased intracellular FPP level through metabolic engineering. A MEP pathway has been shown to synthesize IPP and DMAPP efficiently in E. coli. Because of its high hydrophobicity and low volatility, decane was chosen to extract and solubilize FOH from culture broth. The decane overlay in the two-phase culture did not affect growth, and FOH could be solubilized in the decane phase with negligible volatile loss. We adopt 1 mL of decane overlaid to 5 mL of culture broth. Two-phase culture of E. coli JM109 (BBa_K1653025) was carried out in 2YT medium containing 1% glycerol at 29°C for 48 h. The decane phase of the two-phase culture was collected to analyze the FOH content by GC-MS. In the GC-MS analysis (Fig. 4A-G), there was a main peak at 8.5 min in the collected decane phase sample, which corresponded to the reference solution of the standard FOH compound dissolved in decane. Mass spectrometry confirmed that the peak at 8.5 min was FOH (Fig. 4-A). However, the peak was not observed in two-phase culture without introducing BBa_K165025. The formation of FOH from FPP was further confirmed by blocking FPP synthesis. In the GC-MS, the FOH peak was observed in E. coli JM109 (BBa_K1653025) culture, whereas no peak was observed with transformed E. coli JM109. It was found that FOH need not only ispA(BBa_K1653018) but also MEP(BBa_K1653024) in E. coli. We submit new part(BBa_K1653025) as producing FOH.
Gas Chromatography/Mass(GC/MS)

Geraniol (GOH) production device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653027 BBa_K1653027])
To realize "Flovorator" we need to make E. coli produce Farnesol. Surprisingly, We may have succeeded in farnesol production by geraniol production device.
Our new biobrick device "Geraniol production device" worked as expected. Because we validate this fact by questionnaire survey analysis experimentally. We have documented and submitted this new Biobrick device([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653027 BBa_K1653027]) to iGEM Registry .
![]()
GOH is generated through GPP hydrolysis by geraniol synthase. A MEP pathway has been shown to synthesize IPP and DMAPP efficiently in E. coli. E. coli engineered with geraniol production device ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653027 BBa_K1653027]) showed different smell as compared with counterpart control (pSB1C3) and WT. This result derived from Questionnaire survey. And then we tried to detect that the geraniol generated by engineered "E. coli" by GC and GC-MS. However, the GOH were not detected. These result may indicate that E. coli with GOH production device produce smaller amouts of than can be detected by GC and GC-MS.
-
 fig9:The results of the questionnaire survey WT was 20% of the total, and recombinant(JM109/GES) was 80% of the total, suggesting that recombinant(JM109/GES) was stronger smell than WT. p-value is assumed fifty‐fifty that normally can occur, and the difference compared to that assumption. In this result, p-value is less than 0.01. This probability is beyond the range that can occur by chance. This experiment indicate that recombinant(JM109/GES) might being synthesize geraniol.
fig9:The results of the questionnaire survey WT was 20% of the total, and recombinant(JM109/GES) was 80% of the total, suggesting that recombinant(JM109/GES) was stronger smell than WT. p-value is assumed fifty‐fifty that normally can occur, and the difference compared to that assumption. In this result, p-value is less than 0.01. This probability is beyond the range that can occur by chance. This experiment indicate that recombinant(JM109/GES) might being synthesize geraniol. -
 fig10:The results of the questionnaire survey recombinant(JM109/Empty vector) was 50% of the total, and recombinant(JM109/GES) was 150% of the total, suggesting that recombinant(JM109/GES) was stronger smell than recombinant(JM109/Empty vector) . p-value is assumed fifty‐fifty that normally can occur, and the difference compared to that assumption. In this result, p-value is less than 0.05. This probability is beyond the range that can occur by chance. This experiment indicate that recombinant(JM109/GES) might being synthesize geraniol.
fig10:The results of the questionnaire survey recombinant(JM109/Empty vector) was 50% of the total, and recombinant(JM109/GES) was 150% of the total, suggesting that recombinant(JM109/GES) was stronger smell than recombinant(JM109/Empty vector) . p-value is assumed fifty‐fifty that normally can occur, and the difference compared to that assumption. In this result, p-value is less than 0.05. This probability is beyond the range that can occur by chance. This experiment indicate that recombinant(JM109/GES) might being synthesize geraniol.
Gold
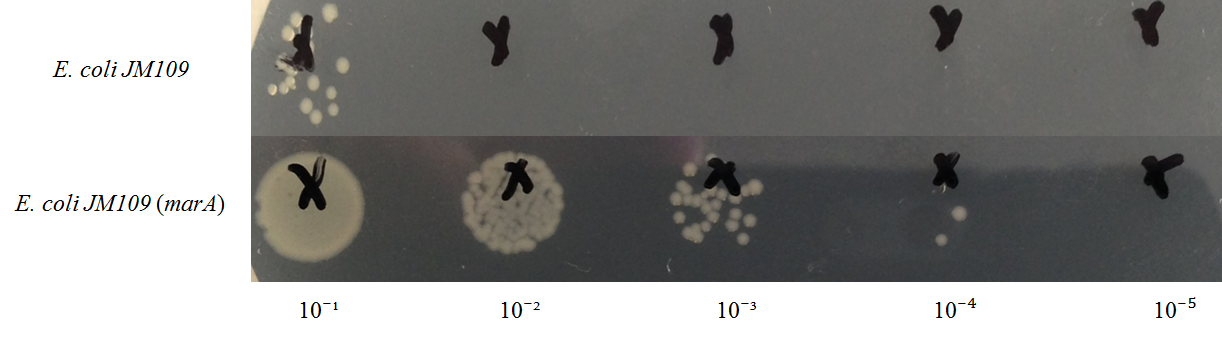
marA :The activator of AcrAB-TolC multidrug efflux pump exports some terpenes
We improved the characterization of a previously existing BioBrick Part [http://parts.igem.org/Part:BBa_K1230000 BBa_K1230000] and submitted this improved BioBrick marA gene as [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653006 BBa_K1653006] to iGEM Registry.
In exsisting part's information of marA, it gives E. coli resistance against kanamycin only. In this year, we confilmed that overepressing of marA gives E. coli resistance against geraniol as one of the terpene and decrease its intracellular concentration. This information is very beneficial for other iGEMers to production of toxic organic substances that are produced using bacteria.
We introduce an activator gene of AcrAB-TolC efflux pump (MarA) to release the geraniol from the cells and increase the content in the media that shows increase these flavors in the "Flavolator". In our study, we confilmed that overexpressing of marA gives host E. coli high resistance against geraniol and reduce intracellular geraniol concentration.
Efficient export of geraniol from E. coli to the mediat
 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653020 BBa_K1653020]
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653020 BBa_K1653020]
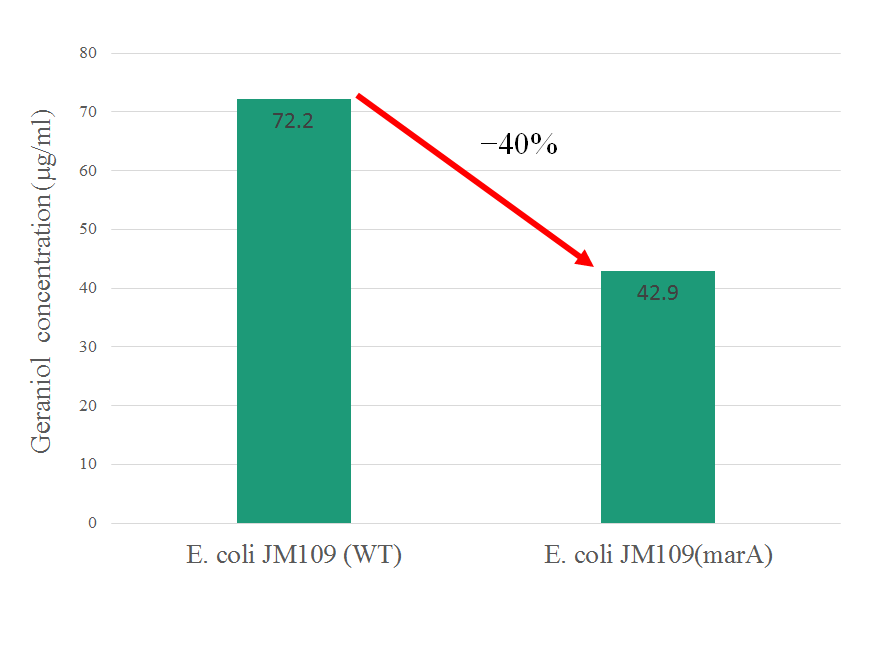
In this figure, intracellular content of geraniol was less in the strain E. coli JM109 (marA) than the strain E. coli JM109 (WT). The concentrations of intracellular geraniol from E. coli JM109 (marA) was 42.9 μg/ml, which was 40% lower than that from of E. coli JM109 (WT), 72.2 μg/ml. This figure is suggesting that internalized geraniol could be more efficiently exported through AcrAB-TolC efflux pump following the presumed activation of this gene by introducing the activator marA gene.
Enhancement of geraniol resistance
 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653020 BBa_K1653020]
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1653020 BBa_K1653020]