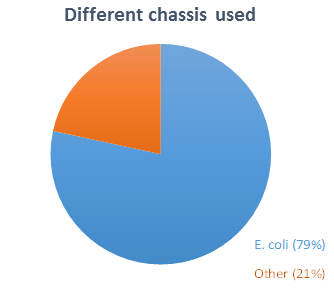
We wanted to design a chassis that could be used as broadly as possible in iGEM, so we asked in our survey to iGEM teams which organism they chose as chassis. Among the 37 teams (representing 12% of all teams) that answered our
and with one or more other chassis). Eight teams worked with other chassis (
, HEK293…).
The main goal of our project is to prevent accidental dissemination in the environment of GEO, and more specifically, of Escherichia coli, the most widely used chassis in iGEM. But before elaborating sophisticated physical and biological containment systems to prevent dissemination, we thought it would be interesting to study the E. coli survival in different natural environments.
Medium
To count our E. coli strains, we chose to use the Mac-Conkey medium. This medium is very selective for Gram negative enteric bacilli, so Escherichia coli. It contains bile salt and crystal violet dye to inhibit Gram positive strains, and neutral red dye which turns pink all colonies able to fermentate the carbon source (for the first test it was lactose, and after sorbitol).
Thus, by adding the appropriate antibiotic to this medium, we thought we would be able to only select our strains, although we needed to check this hypothesis.
Preliminary Study
Our experiment relies on the hypothesis that we will be able to detect selectively our E. coli strain when plating a sample taken from our tested environments on MacConkey plates supplemented with the appropriate antibiotic. We therefore needed to test this hypothesis and verify that the natural microorganisms found in our environment samples would grow or not on our selective plates. We also decided to grow our samples on LB medium as this medium is very rich and allows the growth of many microorganisms. We wanted to see if there was a difference with the MacConkey (MCK) medium, and if we could see the growth of the natural "inhabitants" of our samples.
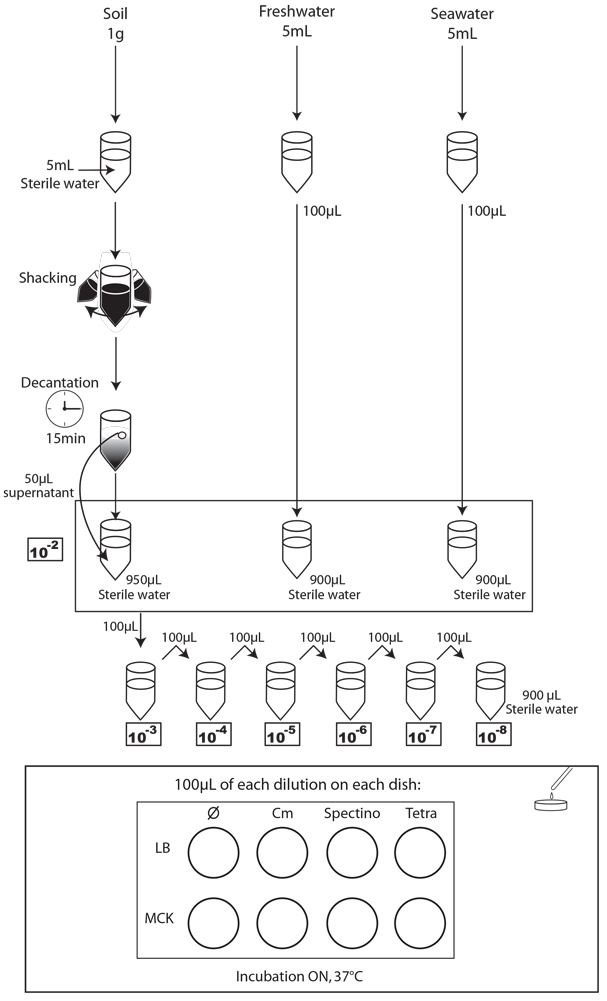
We chose to analyze the soil, seawater and freshwater samples. We prepared 5 dilutions (10-2 to 10-6) of a sample of our natural environment and plated 100 µl on the following media:
- LB
- LB + Spectinomycin (Spectino : 100ng/μL)
- LB + Tetracycline (Tetra : 10ng/μL)
- LB + Chloramphenicol (Cm : 20ng/μL)
- MCK
- MCK + Spectinomycin (Spectino : 100ng/μL)
- MCK + Tetracyclin (Tetra : 10ng/μL)
- MCK + Chloramphenicol (Cm : 20ng/μL)
We next incubated the plates overnight at 37°C and observed them the following day.
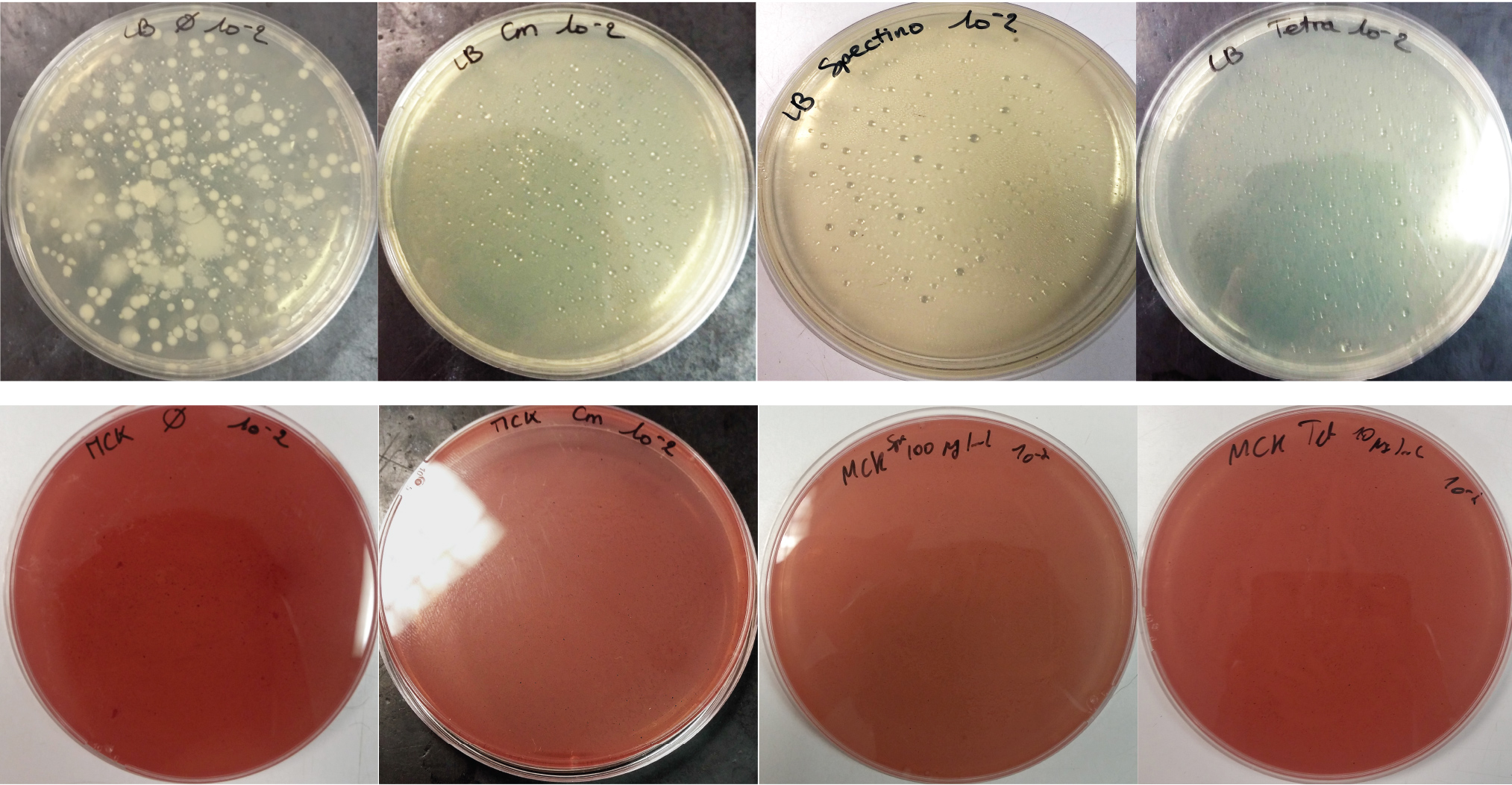
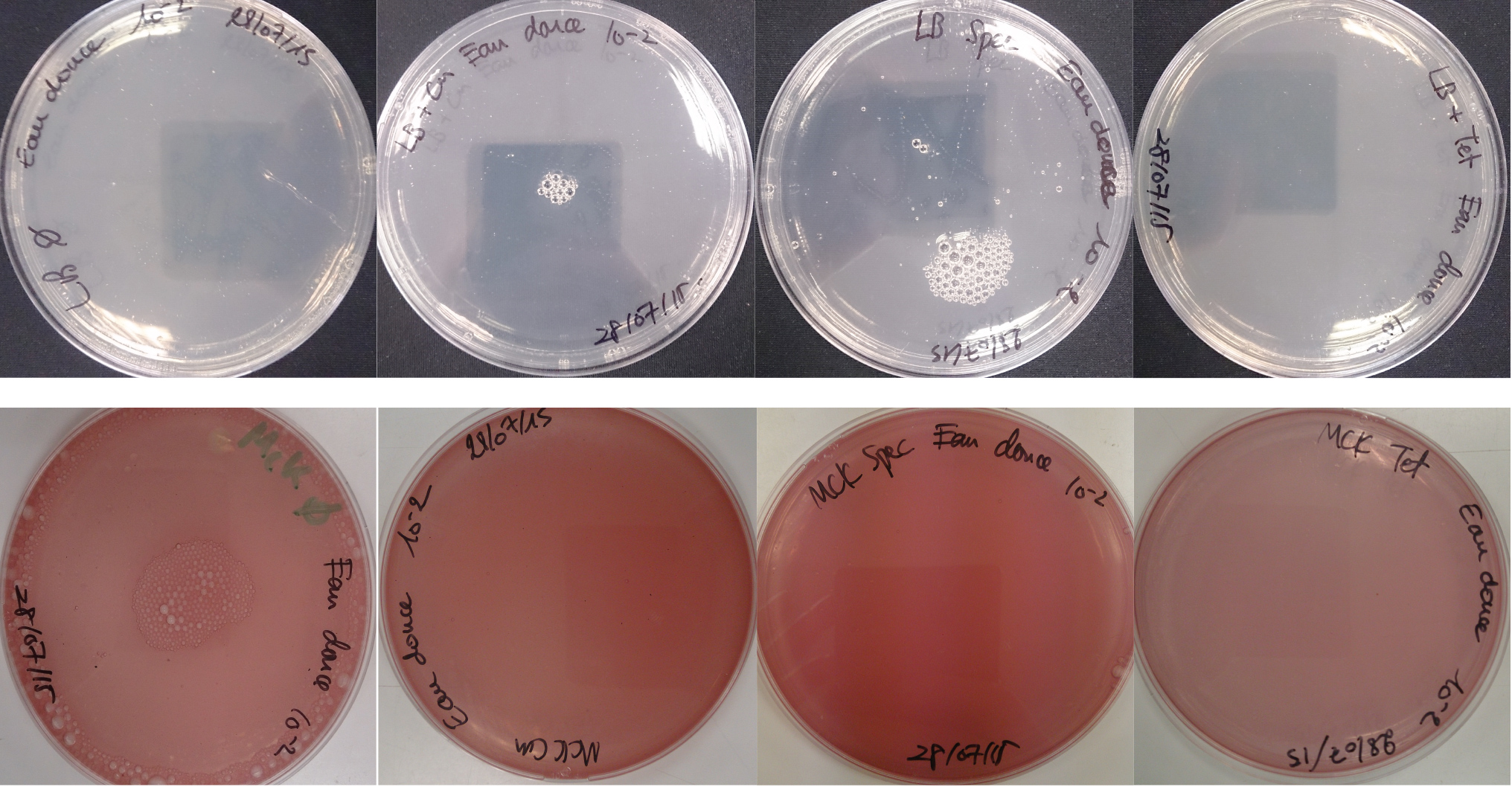
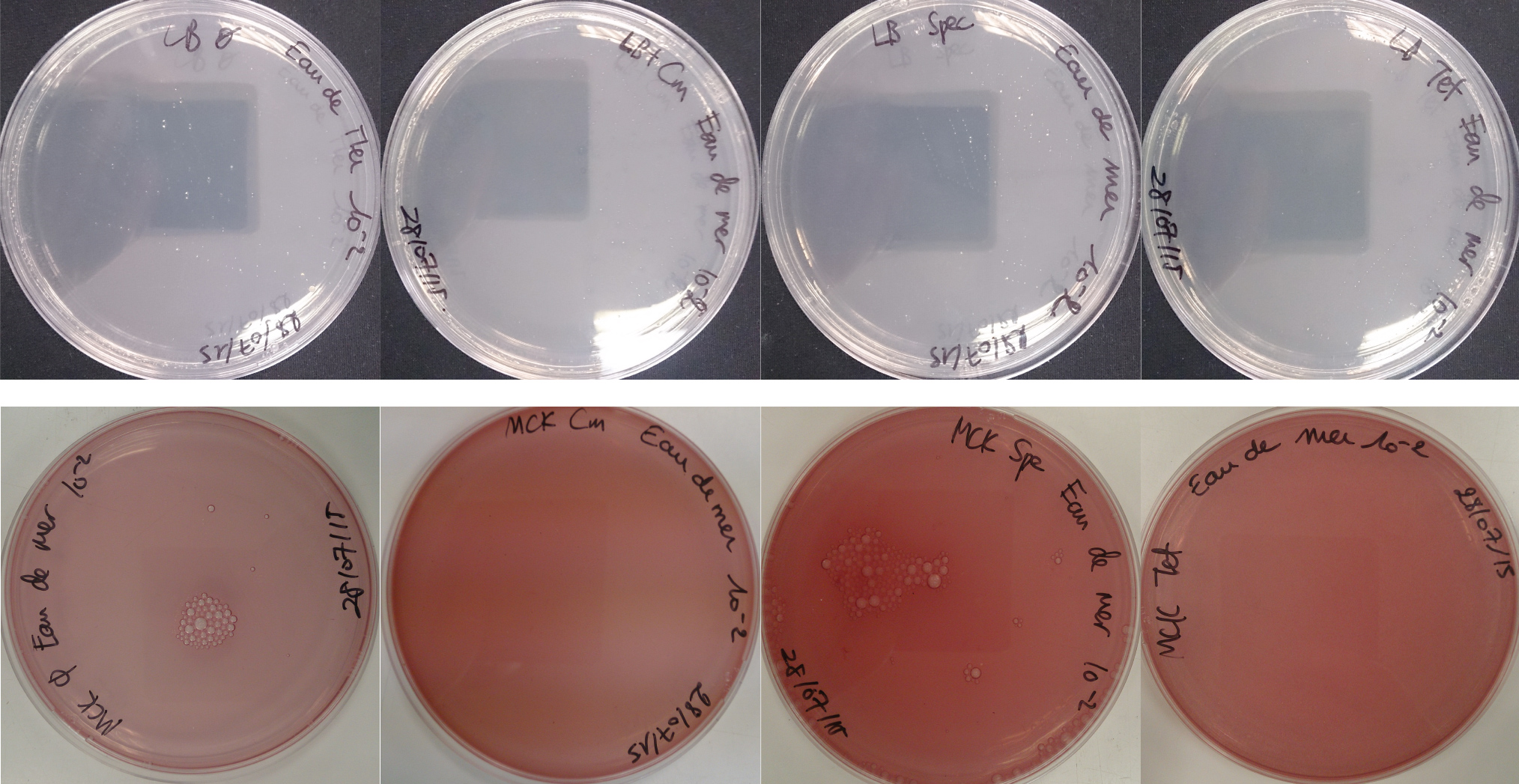
For the soil experiment Figure 2.1, we observed the growth of microorganisms on LB medium for the 10-2 dilution. These micro-organisms, however, appear to be sensitive to three antibiotics tested (chloramphenicol, spectinomycin and tetracycline). They are also unable to grow on the MacConkey medium. No growth of micro-organisms was observed when samples of freshwater Figure 2.2 or sea water Figure 2.3 were plated on either LB or MacConkey media. This may have been expected for the seawater medium, as micro-organisms from this environment may require high salt media for growth.
protocol pre-test survival of E. coli
On the whole, we did not observe growth of “natural” microorganisms on the MacConkey + antibiotics plates, demonstrating that in the conditions tested, the only micro-organisms that we should observed are our specific lab bacteria strains.
Figure 2.1 - Preliminary study for soil environment - Test of soil natural microorganisms growth on LB or MCK with or without Chloramphenicol, Spectinomycin and Tetracycline. Soil samples diluted 10-2
Figure 2.2 - Preliminary study for freshwater environment - Test of freshwater natural micro-organisms growth on LB or MCK with or without Chloramphenicol, Spectinomycin and Tetracycline. Freshwater samples diluted 10-1
Figure 2.3 - Preliminary study for seawater environment - Test of seawater natural micro-organisms growth on LB or MCK with or without Chloramphenicol, Spectinomycin and Tetracycline. Seawater samples diluted 10-1
Study
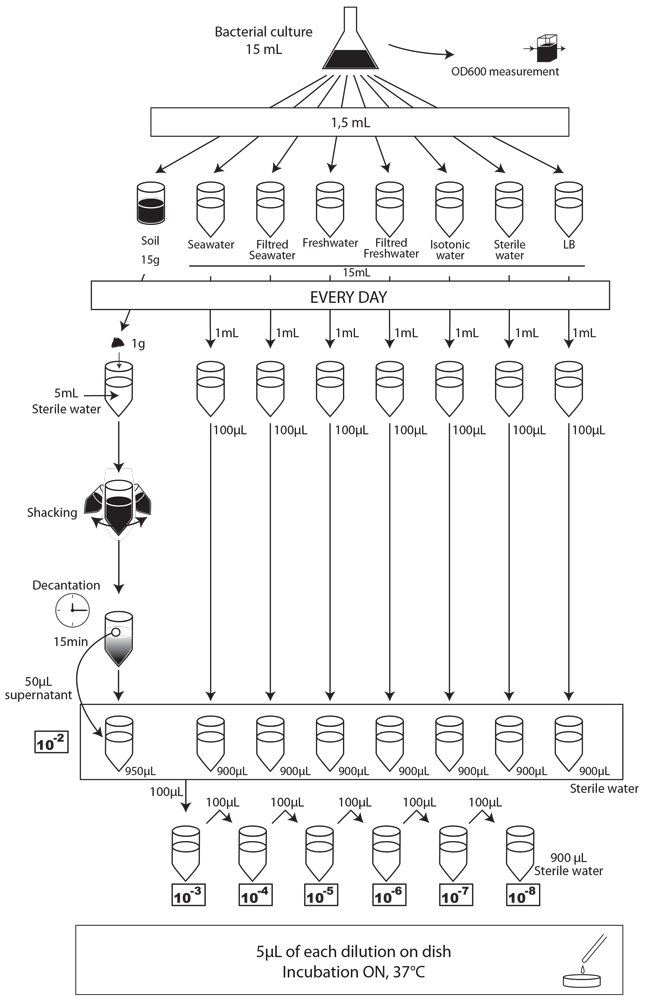
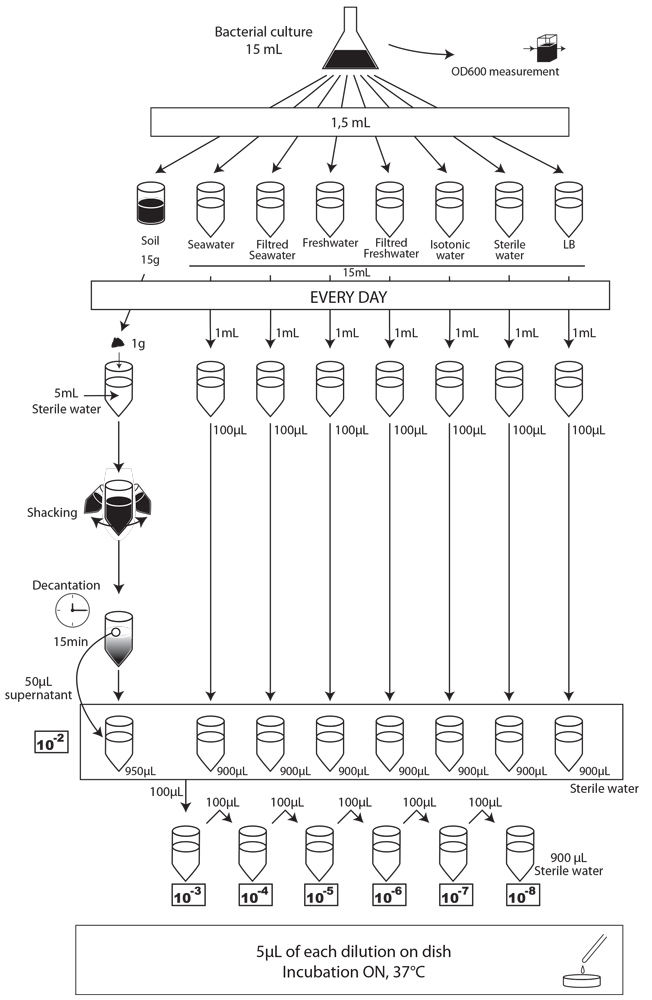
To simulate an environmental contamination by GEOs, we introduced a bacterial culture grown in LB without any antibiotic in our environment samples. Indeed, in case of an accidental dissemination, what could happen to our strain if we spill a bacterial culture into the wild? We chose to use the same protocol for each environment, except an additional step of water dilution for the soil sample.
Protocol experiment : survival of E. coli:
- We prepared a 15mL culture of our three different strains (E. coli 1320, 1693 and 1696), in LB medium, without antibiotic the day before starting the experiment.
- The first day (D0), we measured the optical density at 600 nm (OD600) to evaluate “concentration” in bacteria , and we plated on MCK with the appropriate antibiotic a sample of each culture to count the CFU.
- We put then 1,5mL of each of the three cultures in each of the eight experimental environments (15mL for liquid environment, 15g for soil), and shake it to make a homogeneous mix. Negative controls (-) are environmental samples without culture contamination. All experimental samples were left on the bench, at room temperature, in a 50mL closed falcon tube.
- Each day, we took a 1mL sample of each contaminated liquid experimental environment and prepared dilutions of this sample from 10-1 to 10-8 in isotonic water. For the soil environment, we took 1g of soil, which we diluted in 5mL of sterile water, and after one step of shaking and of 15 min decantation, we pipetted 50 µL of the supernatant and diluted it in 950 µL of isotonic water. This allowed us to obtain the 10-2 dilution. We prepared then the 10-3 to 10-9 dilutions as described for the liquid environments.
- We spotted 5 µL of each dilution on the corresponding MCK plate + antibiotic (no antibiotic for the control).
- We incubated plates overnight at 37°C.
- The next day, we counted the CFU to estimate our strain survival.
protocol experiment : survival of E. coli
Results
It took us time to develop a proper protocol to follow the survival of our strains in the various environments. It was a trial and error process and we had to adjust the protocol after each trial.
Preliminary - experiment
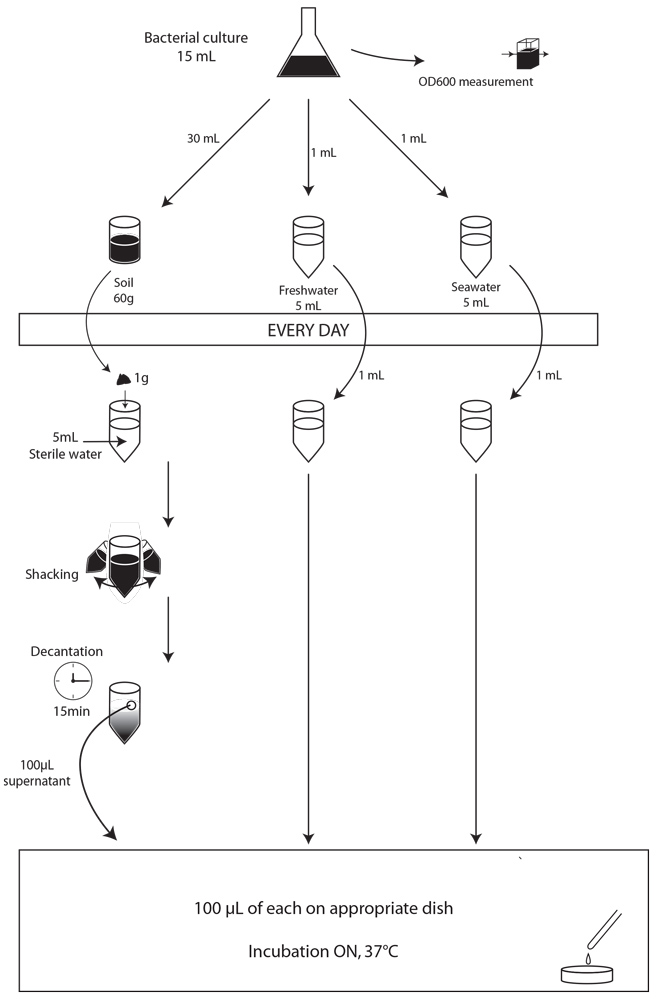
First of all, we made a small-scale preliminary experiment during three days to determine whether our strains were able to survive in our three major environments: soil, freshwater and saltwater, and to determine what would be the optimal concentration of bacteria in our contaminating culture for our experiment. We used 5g of soil, and 5mL of freshwater or salt water. We contaminated them with 1mL of overnight bacterial culture not diluted or diluted 10 or 100 times. For these preliminary experiments, the samples to be plated were prepared as described above, but rather than spotting 5 µl on plates, we plated 100 µl of the dilutions on the adequate MCK plates and incubated the plates at 37°C overnight. Unfortunately, because of some problems with the medium, the results for the experiments with freshwater and saltwater could not be interpreted. The results for the soil experiment are presented in the table 1
On the whole, we observed that after three days of contaminations only, there were no or very few surviving E. coli bacteria in the soil when we diluted our contaminated E. coli culture 10 or 100 times. Moreover, the same decrease trend is observed for each of the three E. coli strains tested, suggesting that the integration of each of the three resistance genes in the genome of the E. coli MG1655 strain is not introducing a bias in our experiment. From this experiment, we concluded that it was more interesting for us to use an undiluted bacterial culture to contaminate our environment in our following experiments.
table 1
|
D0 |
D1 |
D2 |
D3 |
| CM 1 |
more than 1000 |
186 |
111 |
141 |
| CM0,1 |
1744 |
35 |
13 |
2 |
| CM 0,01 |
272 |
1 |
0 |
0 |
| | | | |
| Spe 1 |
more than 1000 |
490 |
424 |
276 |
| Spe 0,1 |
more than 1000 |
67 |
75 |
88 |
| Spe 0,01 |
441 |
0 |
1 |
2 |
| | | | |
| Tet 1 |
more than 1000 |
1308 |
524 |
416 |
| Tet 0,1 |
more than 1000 |
196 |
35 |
62 |
| Tet 0,01 |
414 |
0 |
0 |
0 |
| | | | |
| control |
0 |
1 |
3 |
4 |
First Experiment
We used the same protocol than before, but with 60g of soil and 5mL of water. We made this experiment in triplicate.
Soil
We started a new experiment with more soil (60g) to analyze the contamination evolution during a longer period of time. We planned to do it for a few weeks but after three days, we observed contaminations on our negative control. After multiple trials, we concluded that this was due to the MCK medium used. Thus we changed the MCK stock and did tests from which we concluded that adding sorbitol to the MCK was more selective than adding lactose. Consequently, we decided to start another time the experiment for all the different contaminated media using Sorbitol-MCK rather than lactose-MCK.
Seawater and Freshwater
The first time we put our strains in 5mL of water but we encountered two main problems. The first one was that it was very difficult to spread the 100µL sample on plates. The samples seemed to slide on the medium. And the next day, we can only observe some “lines” of colonies. The second problem appeared on the second day: the medium, normally red, turned into a yellow color. This was probably due to a change of pH in the water sample, most likely due to the metabolism of some microorganisms when adding LB in the environment.
Final experiment
After those pre-tests, we finally had established a protocol that should allow us to carry out our experiment. We chose to test 15g (soil) or 15mL (seawater and freshwater) of our samples contaminated by 1,5mL of our bacterial cultures. We used the following protocol.
We performed CFU analyses each day by depositing 5µL spots of contaminated media on specific MCK plates. The results are presented bellow.
Controls
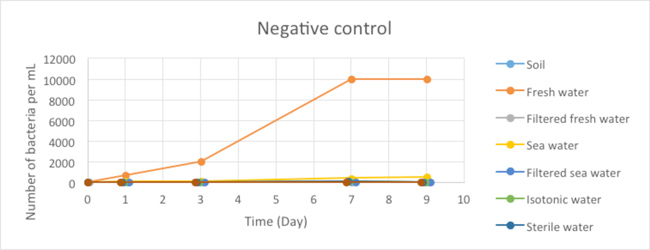
Figure 2.1 - Negative Control
Due to the lack of time, we were not able to conduct these experiments in triplicates and for a longer period of time, which would be required to obtain solid results from which conclusions can be drawn. Nonetheless, these experiments allowed us to formulate some preliminary conclusions. First, in our negative control (corresponding to samples of uncontaminated environments plated on MCK plates without antibiotics), we observed a very limited number of bacteria in all our samples except for the freshwater sample. This number also appeared to be stable in time, suggesting that the population of the micro-organisms that grew on our plates remained stable during the time of our experiment.The important bacterial growth observed for the non-filtrated freshwater is surprising and inconsistent with the results obtained for the other samples. It will be necessary to repeat this experiment to check whether this is observed reproducibly.
Before interpreting the results of our contamination experiments, it is important to remember that we did not observed the growth of any micro-organisms from the various environments tested when antibiotics were added to the MacConkey medium.
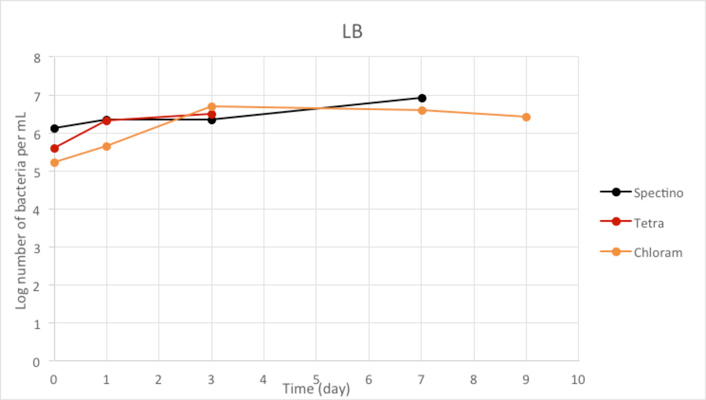
Figure 2.2 - Positive Control
We used LB medium for our positive control. We can see that during the first few days, the E. coli population is growing, before stabilizing. It should be noticed that for Day 9, we have only one measure, because of manipulation errors (wrong dilutions) with the samples LB+ PhB1393 and LB+PhB1396.
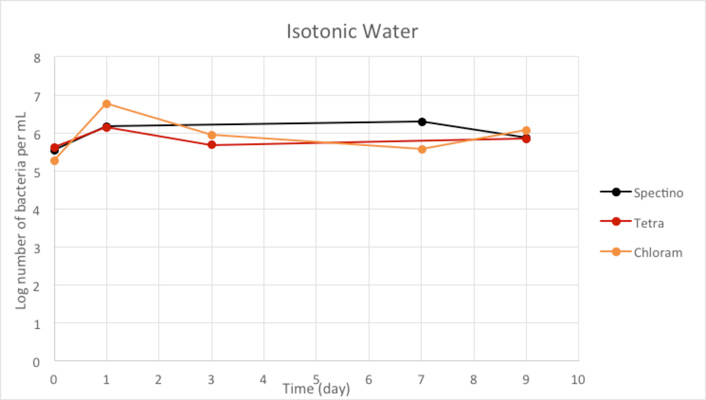
Figure 2.3 - Isotonic water
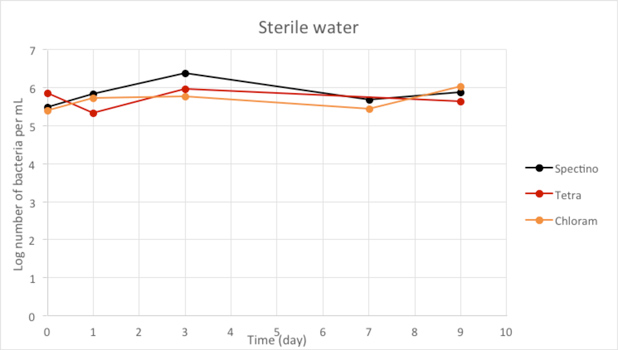
Figure 2.4 - Sterile water
Our controls of E. coli strains in isotonic water or sterile water shows that they survive perfectly well in these environments.
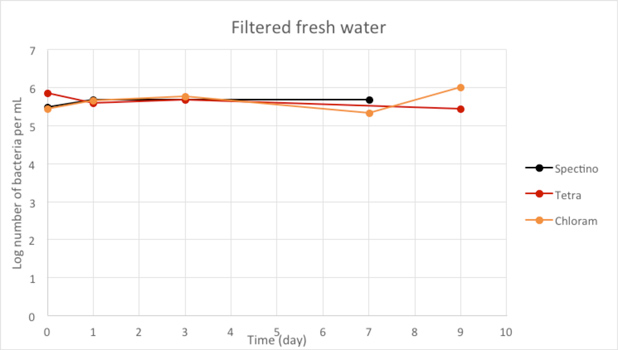
Figure 2.5 - Filtered fresh water
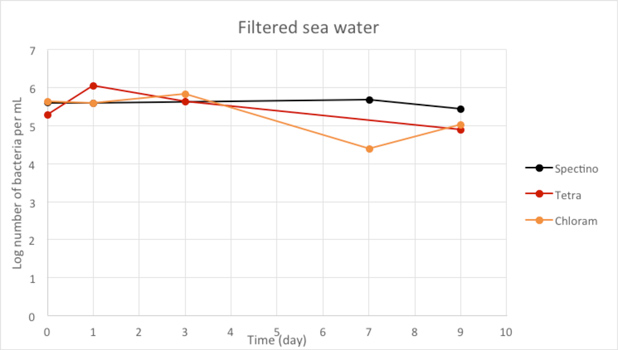
Figure 2.6 - Filtered sea water
When we filtered our water samples, we eliminated all the micro-organisms present in these environments. We observed that on the whole, the number of laboratory E. coli bacteria in these two environments remain stable for the length of the experiment.
Experiment
All the environmental samples contained various organisms (as demonstrated by the pre-test), so these three experiments are close to the environmental conditions (as the temperature or light fluctuation).
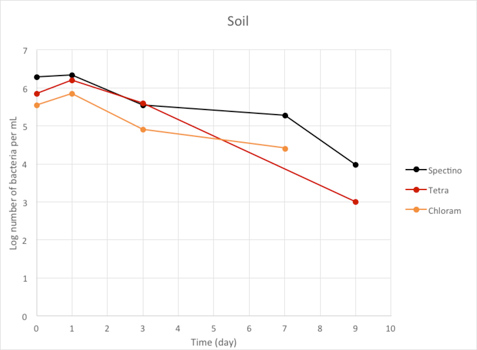
Figure 2.7 - Soil
We observed a large decrease of the number of living laboratory E. coli bacteria with time (100 to 1000 fold decrease between day 1 and day 9), for the three tested strains.
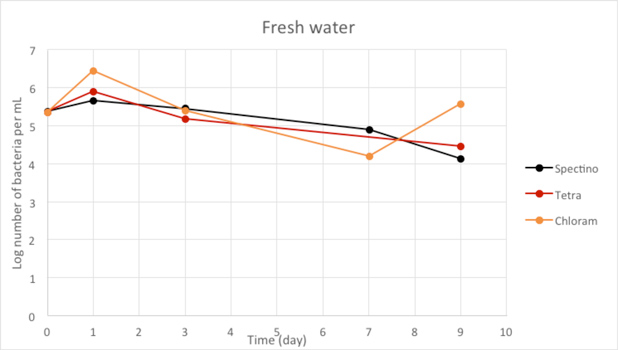
Figure 2.8 - Fresh water
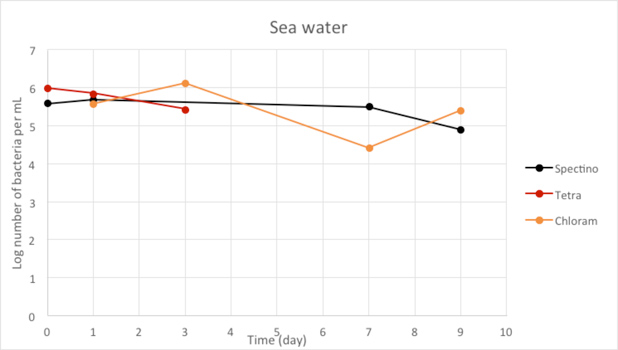
Figure 2.9 - Sea water
For the freshwater and seawater environments, we observed a small decrease or a stability of the population of living laboratory E. coli bacteria with time. The results for the seawater environment is surprising, as it is has often be described that E. coli populations rapidly decline in this environment, although in some experiments, a lag time before the decline of the population of up to 20 days has been observed. (Vaccaro et al., Am J Public Health Nations Health. 1950 Oct; 40(10): 1257–1266).
Conclusions
The experiments we conducted showed that during at least 9 days, laboratory E. coli bacteria can survive in environments as varied as the soil, the water of a pond or ocean water. We were careful in designing our experiments to try to limit as much as possible the factors that could bias the results of our study (three E. coli strains tested) and included all the control we could think of. We also noticed that in all the liquid conditions tested, a biofilm was visible from day 3. The fact that these biofilms appeared in conditions such as sterile water or filtered water led us to the conclusion that our E. coli strains can form biofilms, a property that is thought to be important for the survival in the environment (Vogeleer P et al., Frontiers in Microbiology, vol5, 2014).
We did not study here if DNA fragments could be exchanged between organisms, which could be a real issue, especially in case of a prolonged survival in the environment. Altogether, this experiment confirmed the necessity of elaborating physical and biological containments for GEO that could be released in the environment, either accidentally or for a specific application.
A special thank to Purificación López-García's team to explain the limit of our experiment and help in the process of making it. We also thanks the Philippe Bouloc's team (Signaling and Regulatory Networks in Bacteria) for the MCK and the help.
The Temperature-Based System for E. coli: SafetE.coli
Overview
To protect the environment from a possible Genetically Engineered Bacteria (GEB) contamination, we created a two-level protection system. First a biological containment was built, which could be improved with the addition of a physical containment system level depending applications. We choose to build a system which the E. coli viability was restricted to a narrow range of temperatures. Such system should then be combined with the physical containment system.
Problematic
Nowadays, a large number of thermosensitive systems are commonly used in the biology field. Two points appeared to be important when we started to design our device: first, it must be durably functional in time to avoid the spread of GEB. Secondly, the system must be flexible and adaptable to the users’ needs.
Details
Why our device is safe and stable in time
In order to create a safe system, we choose to control the survival of bacteria by modulating the expression of several essential genes.
The targeted essential genes were chosen in accordance with the literature on the subject. They have to control a vital function in the cell, be well-characterized, have a constitutive expression, and not be redundant in the bacterial chromosome. Taking into account these various parameters, adenylate kinase (adk), alanyl-tRNA synthetase (alaS), DNA polymerase III subunit delta (holB), methionyl-tRNA synthetase (metG), phosphoglycerate kinase (pgk) and tyrosyl-tRNA synthetase (tyrS) should for instance be good candidates. The more of these target genes are controlled by our system, the less likely is the bacterium to live outside the lab.
The aim of our system is to control the expression of these chosen essential genes.
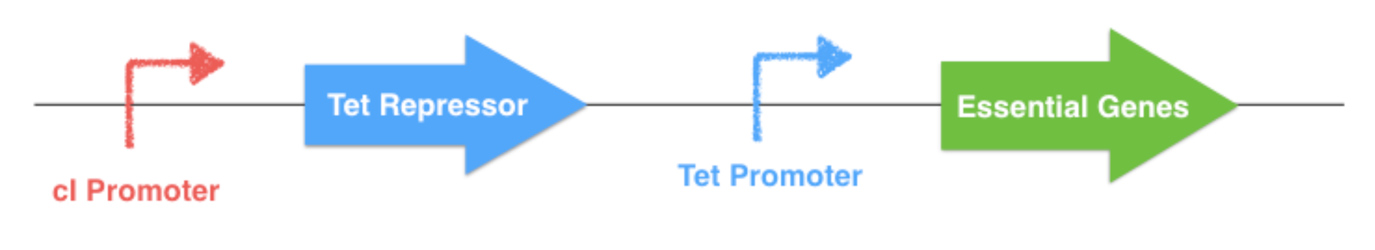
To do this, a replacement of their natural promoter by homologous recombination has to be made. The repressible Tet Promoter, which is controlled by the Tet Repressor, will replace the native gene promoter. The repressor gene with a specifically engineered promoter will also be inserted on the chromosome. For this project, the classical Tet system will be modified by placing theTet Repressor gene under the control of a thermosensible repressor, this repressor being downstream a thermoinducible promoter. Thereby the expression of the essential gene is indirectly determined by the outside temperature.
The thermal control part of the system (described below) could be implemented on a plasmid or onto an other localization of the E. coli chromosome. If this plasmid is lost by the bacterium, the Tet Repressor will constitutively inhibit the transcription of the downstream essential gene, and the bacterium will die. The same scenario will happen if one of the system elements undergone a mutation.
Figure 3.1 - Chromosomal essential genes control - E. coli chromosome
How our system could be adapted to biologists needs
In order to answer the problematic of system adaptability, we choose to use molecules that undergo structural changes depending on the temperature.
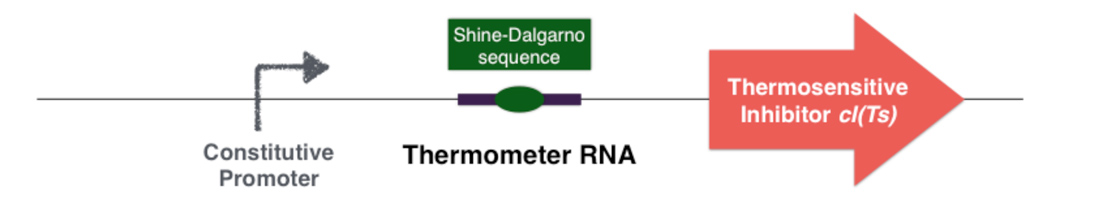
To achieve this goal, two molecules have been selected: an RNA thermosensor and a thermosensitive repressor. These two molecules define a high and a low temperature limitation. They can be introduced in a plasmid or introduced inside of the chromosome, under the control of a constitutive promoter.
Figure 3.2 - Thermal control device - Plasmid or chromosome
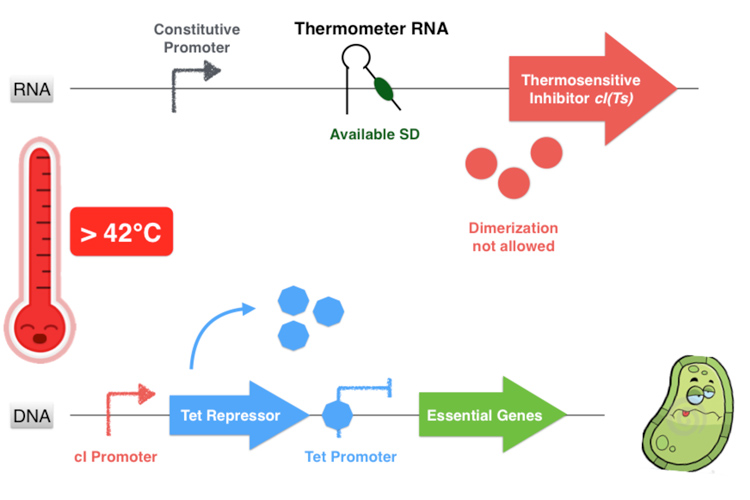
The first part of the system consists in a thermosensitive variant of transcriptional repressor cI from bacteriophage lambda. cI repressor protein acts as a dimer to bind to promoters and repress downstream gene expression. For cI(Ts), a cI protein variant, the dimerization happen only if the temperature is under 42 degrees, since at high temperatures cI(Ts) is denatured and cannot dimerize. Consequently the cI(Ts) protein is not functional above 42 degrees. Other variants of the cI repressor can be created by inducing mutations near the dimerization site, in order to change the denaturation temperature.
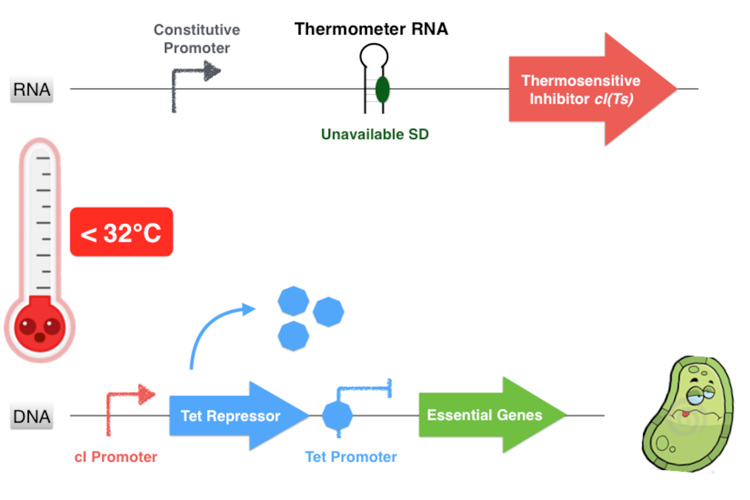
Behind the thermosensitive cI variant, instead of its native RBS, we used an RNA thermometer which is based on a ROSE RNA thermometer retrieved from Bradyrhizobium japonicum. The temperature sensitive hairpin of this RNA thermometer that contains the Shine Dalgarno (SD) sequence is closed when the temperature is under 32°C, that structurally prevent the ribosome binding to the downstream RNA sequence, and so it cannot start to translate RNA. The translation can only be initiated when the temperature is around 32°C, when the hairpin is opened and so the SD sequence is available for the ribosome binding. The switch temperature of the thermometer RNA is easily tunable by mutations in order to stabilize or destabilize its Shine Dalgarno hairpin.
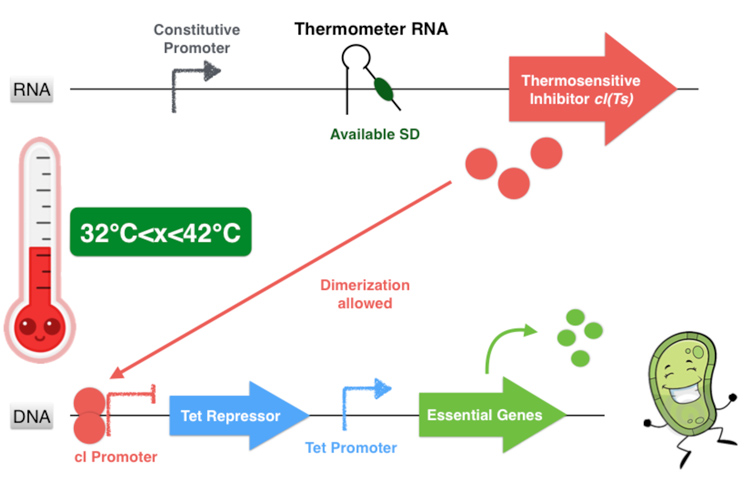
The global thermal system functioning
Overall, our device consists in an inhibition of an inhibitor to allow the essential genes expression. This situation happens only if the environment temperature is comprised between 32 and 42 degrees because it is the only range of temperature that allows the production of a functional dimer of repressor cI. Indeed, from 32°C the thermometer RNA allows the ribosome to translate the RNA. And secondly, with a temperature below 42°C, the cI protein is well-folded and can form a homodimer. This cI pair then binds to the cI promoter, thus inhibiting the Tet Repressor and allowing thereby the expression of the essential genes under its control. Finally, at temperatures comprised between 32 and 42°C, our system allows the production of the proteins essential for the bacterium’s survival.
Figure 3.3- First scenario : Permissive temperature range
On the contrary, if the temperature is under 32°C, the hairpin of the thermometer RNA is hybridized on itself and masks the SD sequence, inhibiting the translation of the cI protein. Thereby the Tet Repressor is constitutively produced and inhibits the essential genes expression, leading to the bacterium’s death.
Figure 3.4- Second scenario : Lower non-permissive temperature range
If the temperature is over of 42°C, even if the thermometer RNA hairpin is linearized and allows the ribosome reading of the cI RNA sequence, the cI inhibitor cannot form homodimers and is not functional anymore. Thus the Tet Repressor is also produced and inhibits the essential protein production leading to the bacterium’s death.
Figure 3.5 - Third scenario : Higher non-permissive temperature range
Result
In order to design a safety E. coli chassis, numerous test constructs have been made. Each of it is intended to check the functionality of one part of our final system. [[File:https://static.igem.org/mediawiki/2015/f/fe/Paris_Saclay-Thermal_R1.jpg|400px|center]] Figure 3.6 – Final system on E.coli chromosome
To functionally characterize our parts, a fluorescent reporter: a Cyan Fluorescent Protein with a ssRA tag has been introduced downstream, to permit a fast destruction of the reporter and so visualize the fluorescent switch rapidly.
Finally 38 BB have been constructed, but due to a lack of time, only 2 biobricks have been properly characterized by fluorescence microscopy: the BBa_K1707026 which correspond to the ssRA tag test and the BBa_K1707028 which is a cI promoter test.
Figure 3.7 – System components test
First, to check the efficacy of the ssRA tag to destabilize the CFP, the intensity of fluorescence of two BB have been confronted: BBa_I13600 (Figure 3.9) and BBa_I13602 (Figure 3.10). Both are CFP under control of a Tet promoter, but only the BBa_I13602 has a ssRA tag. Through fluorescence microscopy we can see that the bacteria transformed with CFP (BBa_I13600) are more fluorescent than the bacteria transformed with CFP-ssRA (BBa_I13602). Our negative control, DH5α bacteria (Figure 3.8 ) don’t show significant fluorescence. In conclusion the ssRA tag seems to permit a destabilization of the CFP protein, reducing its lifetime. This experiments allowed the first characterization of the BBa_I13602.
Figure 3.8 – DH5α microscopy observation (Negative control)
A) Differential interference contrast
B) excitation with led 437 nm/17nm (CFP), 200 ms of exposure
Figure 3.9 – BBa_I13600 microscopy observation (Positive control)
A) Differential interference contrast
B) excitation with led 437 nm/17nm (CFP), 100 ms of exposure
Figure 3.10 – BBa_I13602 microscopy observation (Positive control)
A) Differential interference contrast
B) excitation with led 437 nm/17nm (CFP), 100 ms of exposure
In order to test the strength of two other promoters used in our system, a CFP-ssRA has been introduced downstream a constitutive promoter (BBa_K1707026 Figure 3.11) and a cI promoter (BBa_K1707028 Figure 3.12). Under fluorescence microcopy, the intensity of fluorescence visualized for these two new BB seems to be equivalent to the results obtained with the BBa_I13602. In conclusion, we observed a minor intensity of fluorescence for these two strong promoters with a ssRA tagged CFP, in comparison to a medium promoter (Ptet promoter) with classic CFP (see Figure 3.9).
Figure 3.11 – BBa_K1707026 characterization
A) Differential interference contrast
B) excitation with led 437 nm/17nm (CFP), 100 ms of exposure
Figure 3.12 – BBa_K1707028 characterization
A) Differential interference contrast
B) excitation with led 437 nm/17nm (CFP), 100 ms of exposure
Conclusion and perspectives
To conclude, we designed a biosafety system based on thermal control. We could imagine tailoring this system for different temperature ranges to meet the user’s needs. The cI repressor could be mutated at different positions to try to modify the transition temperature, as could be the switch temperature of the RNA thermometer. In addition, our project is designed to provide the iGEM community with biobricks that could easily be integrated in the chromosome of any Escherichia coli chassis with a safety system on it. In principle, it could be adapted to other bacterial chassis. However, one biological containment system is likely to be insufficient, and to reduce the likelihood of an environmental contamination, it would be better to multiply the biological (auxotrophic selection, toxin/antitoxin system) or physical (such as the silica beads presented below) security systems. Nevertheless, an increasing number of bacterial modifications may seriously impact the bacterium behavior.
Physical containment
Considering the non-negligible risk of escape of a GEO, we wanted to add an extra layer of safety in our project. Therefore, we devised a way to physically contain E. coli and other bacteria without hindering it to perform its main function. Indeed, the system we designed is a porous glass beam in which bacteria can grow, survive and carry out their function. The nutrients of the external medium (sugars, amino acids, etc...) can penetrate inside the devise but the bacteria cannot escape from it.
Thus, the SafetE.coli chassis can be in a safer manner in contact with natural environments, which is an important aspect when the project competes in iGEM competition tracks such as Environment or Health and Medicine.
Some teams include in their project a devise, more or less sophisticated, in which the GEO or their products will perform its function. Here we can cite the 2014 project of NCTU Formosa where we can found an example of such devise: The Pyramid Trap containing PBAN (Pheromone Biosynthesis Activating Neuropeptide) produced by a Genetically Engineered Escherichia coli. However, other teams do not have the time or means to do as well.
Thus, the 2015 Paris Saclay team project offers to all iGEM teams a fast, efficient and inexpensive way to build an impenetrable shelter for bacteria.1
The system principle
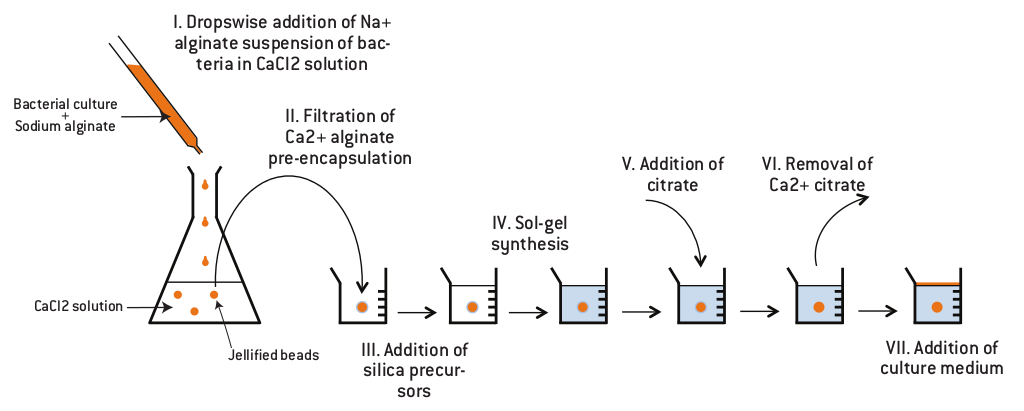
The aim of the containment is to create a physical barrier which should prevent accidental spreading of bacterial culture. The idea is to make a containment where bacteria can grow but which does not allow them to be in contact with the environment. Bacteria will be contained inside silica monoliths. To enable cell growth, we found a protocol which describes how to make cavities inside silica monoliths possible.
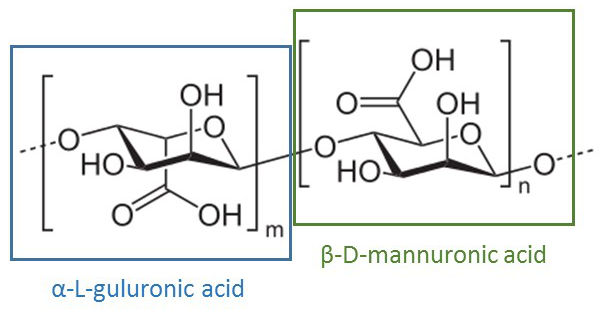
First, cells are encapsulated in an alginate gel. The alginic acid polysaccharide is a linear copolymer of β-D-mannuronic and α-L-guluronic acid extracted from brown algae or bacteria.
Alginic acid polysaccharide
The bacterial culture is mixed with sodium alginate and the solution is dropped in a chloride calcium solution. At neutral pH, carboxylic acid functions are deprotonated so that the polymer bears a global negative charge, usually compensated by sodium ions. Addition of divalent cations such as Ca2+ induces cross-linking of the polymer, and therefore gel formation.
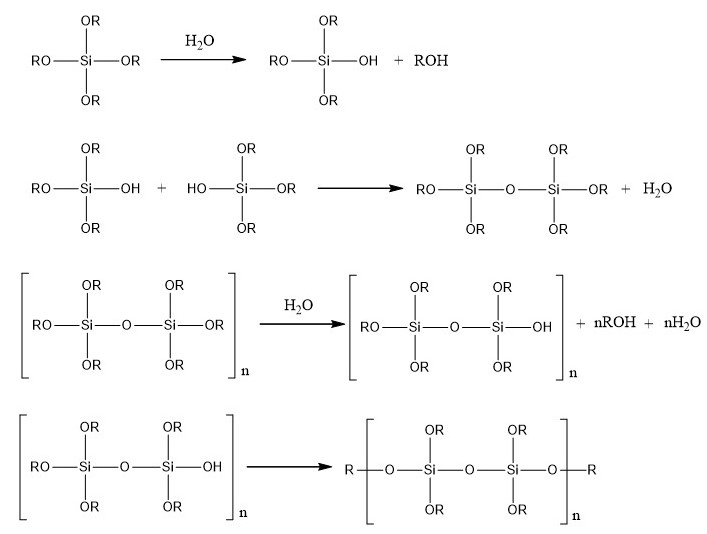
Then, silica monoliths are created around the beads obtained by a sol-gel process which allows the formation of silica gels at room temperature from aqueous precursors (Brinker and Scherrer 1990). The reactions involved in this process are presented in the figure below.
Polymerization reactions involved in the sol-gel process
Finally, the gel is dissolved by using an acid which chelates Ca2+ so that cells have a cavity where they can grow and which can be filled with a defined growth medium.
Protocol
The first one is performed by dropwise addition of a 1.5% (w/w) sodium alginate cells suspension in a 0.1 M CaCl2 solution. After 10 min stirring, about 3 mm diameter beads are easily collected by filtration. The calcium alginate polymer prevents cell contact with synthesis precursors. The second step consists of silicate polymerization in the presence of commercial silica nanoparticles (Ludox HS40 from Aldrich), leading to a nanoporous monolithic structure. Monoliths are prepared at room temperature by mixing 2 volumes of 1.25 M sodium silicate with 1 volume of colloidal silica and 1 volume of succinic acid (5 wt %) into a recipient containing the alginate-cells bead. Once the sol-gel polymerization reaction is completed, the stiff monolith obtained is left in contact with 0.05% potassium citrate during three hours. To provide necessary nutrients to the immobilized cells, potassium citrate solution is further replaced by LB medium according to encapsulated cell strain requirements.1,2
Physical containment protocol
Results
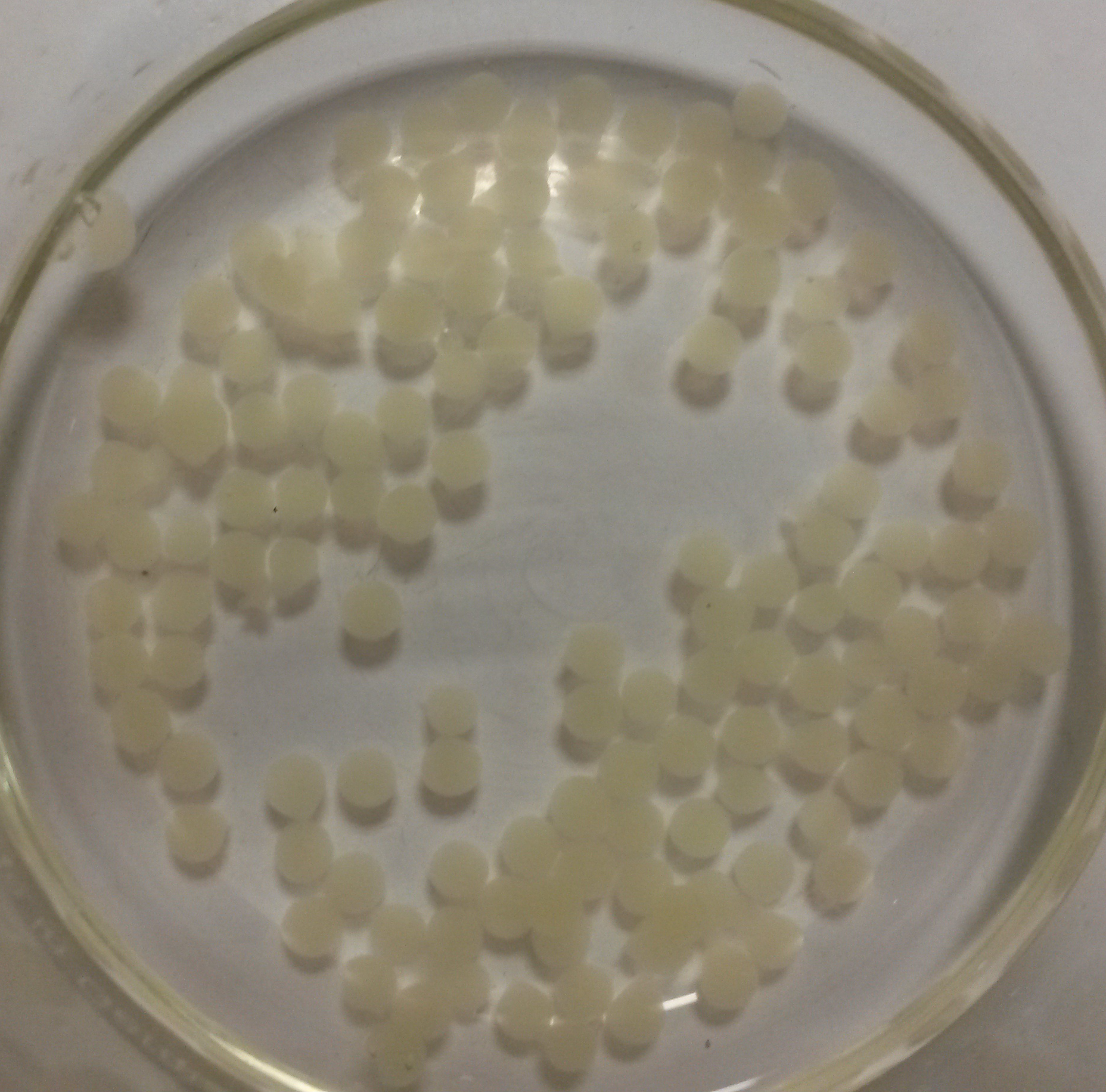
We obtained nice beads which are properly jellified and measure about 3 mm as expected.
Alginate beads of bacterial culture
The silica monolith was obtained only when we let it in contact with air.
Alginate beads with few silicate at their surfaces containing bacterial culture
Conclusion and Perspectives
We have succeeded in creating a physical containment for bacteria which is cheap, fast to build and should prevent unintended contacts between GEOs and the environment. Further experiments are recquired in order to test such system. It will be interesting to see if bacteria do not escape from the silica monolith and if they can survive inside it. The Aachen team suggested us different devices to measure the growth rate. The first one can be used to determine biomass concentration using capacity measurements.3 The second one measures growth rate using an optical detection system that is based on measuring the intensity of back-scattered light from bacterial cells suspended in liquid culture.4
References
(1) Mercedes Perullini, Mat´ıasJobba´gy, Galo J. A. A. Soler-Illia, and Sara A. Bilmes, Chem. Mater. 2005, 17, 3806-3808
(2) Mercedes Perullini, Frédéric Orias, Claude Durrieu, Matias Jobbagy, Sara A. Bilmes, Biotechnology Reports, 2014, 4, 147-150
(3) Noll, T., & Biselli, M., Journal of biotechnology, 1998, 63, 187-198
(4) Toprak, E., Veres, A., Yildiz, S., Pedraza, J. M., Chait, R., Paulsson, J., & Kishony, R., Nature protocols, 2013, 8, 555-567
Perspectives
Modeling