Difference between revisions of "Team:William and Mary/Parts"
Adhalleran (Talk | contribs) |
|||
| Line 461: | Line 461: | ||
action=raw&ctype=text/javascript"></script> | action=raw&ctype=text/javascript"></script> | ||
| − | <script type="text/javascript" src=" | + | <script type="text/javascript" src="https://2015.igem.org/Team:William_and_Mary/modernizerJS?action=raw&ctype=text/javascript?"></script> |
<script type="text/javascript" src="https://2015.igem.org/Template:William_and_Mary/rubickJS? | <script type="text/javascript" src="https://2015.igem.org/Template:William_and_Mary/rubickJS? | ||
action=raw&ctype=text/javascript"></script> | action=raw&ctype=text/javascript"></script> | ||
</html> | </html> | ||
Revision as of 02:57, 19 September 2015
All Parts (By Category) This year we submitted 25 parts to the iGEM registry. Browse them below. Integrator Cassette For our method of genome integration the input is linear DNA, generated by PCR, containing what you would like to integrate onto the genome and an antibiotic resistance cassette to allow for selection. The galK Integrator allows digestion with the standard BioBrick enzymes and 3A assembly of your part of interest to create the integration construct (see right).
This product can then be amplified using primers (details found here) and then used in the integration protocol. We have successfully used the integrator to incorporate stretches of DNA up to 2.1kb into the galK locus, not including the resistance cassette (1179 bp). Antibiotic Operon dCas9s Part BBa_K1795000 is a dCas9 protein-coding region that has been optimized for expression in E. coli. Additionally, we created a functional dCas9 operon (BBa_K1795001) that, when transformed into E. coli is constitutively expressed. For further details, please see our Basic Part page. gRNAs Part:BBa_K17950002 R0010 gRNA Part:BBa_K17950003R0051 gRNA Part:BBa_K17950004R0062 gRNA Part:BBa_K17950005R0011 gRNA Part:BBa_K17950006J23100 gRNA Part:BBa_K17950007J23101 gRNA Part:BBa_K17950008J23106 gRNA Part:BBa_K17950009J23117 gRNA Part:BBa_K17950010J23119 gRNA Part:BBa_K17950011I13453 gRNA Part:BBa_K17950012I0500 gRNA Part:BBa_K17950013Scrambled gRNA To complete our creation of a codon-optimized dCas9 parts, we created functional gRNAs that target the most commonly used promoters in iGEM. These parts (BBa_K1795002- BBa_K1795012), when transformed into E. coli, constitutively express gRNA that, in complex with our dCas9 variant, will repress transcription of the targeted promoter. We have also contributed a Scrambled gRNA (Part:BBa_K17950013) that does not target any region in the E. coli genome and can be used as a negative control. For further details about our gRNAs, please see our Part Collection page.
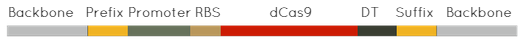
CFPs/YFPs Under Various Promoters Part:BBa_K1795014YFP-LVA under R0040 Part:BBa_K1795015CFP-LVA under R0011 Part:BBa_K1795016YFP-LVA under R0011 Part:BBa_K1795017YFP-LVA under R0062 Part:BBa_K1795018CFP-LVA under I13453 Part:BBa_K1795019YFP-LVA under I13453 Part:BBa_K1795020CFP-LVA under R0051 Part:BBa_K1795021CFP-LVA under I0500 In order to test multiple promoters for their intrinsic noise, we needed to construct composite parts that had the structure seen to the right.
In each construct the RBS and DT were kept constant throughout, but the promoter used to drive transcription was changed. Each construct was made in a CFP and YFP variant. Both the CFP and YFP used in this project have an LVA tag, decreasing the half-life of the protein. All of these parts have been sequenced confirmed and functionally validated. Dual Fluorescent Plasmid This plasmid consists of both a CFP-LVA driven by R0010 and YFP-LVA driven by R0010. This allows future teams to use this part to investigate transcriptional noise either on a low copy number plasmid or by integrating this part using our galK integrator.