Notebook
Contents
- 1 Week 1 (July 6 ~)
- 2 Week 2 (July 13 ~)
- 3 Week 3 (July 20 ~)
- 4 Week 4 (July 27 ~)
- 5 Week 5 (August 3 ~)
- 6 Week 6 (August 10 ~)
- 7 Week 7 (August 17 ~)
- 8 Week 8 (August 24 ~)
- 9 Week 9 (August 31 ~)
- 10 Week 10 (September 7 ~)
Week 1 (July 6 ~)
 Human Practice
Human Practice
We started main HP project this year. We had already did some communication and discussion with other iGEM team. The initial idea is to tackle our project from different fields, such as law aspect. We start to get in touch with some of our experts.
 Construction
Construction
July 7
1. Get promotor PcpcB fragment and Kn (kanamycin) fragment respectively from plasmid Bluescript and PRSF by PCR
| PcpcB (from pBluescript) | Kn (from pRSF) | |
|---|---|---|
| ddwater | 45.5μl | 45.5μl |
| Template | 0.5μl(1ng/50μl) | 0.5μl(1ng/50μl) |
| Primer1(10μM) | 2μl | 2μl |
| Primer2(10μM) | 2μl | 2μl |
| PrimeSTAR Mix | 50μl | 50μl |
| Total volume | 100μl (50μl*2) | 100μl (50μl*2) |
| ×34 cycle | 98℃ 3min | 98℃ 3min |
| 98℃ 10s | 98℃ 10s | |
| 56℃ 5s | 58℃ 5s | |
| 72℃ 3s | 72℃ 5s | |
| 72℃ 3min | 72℃ 3min | |
| 12℃ ∞ | 12℃ ∞ | |
| Final concentration(after purification) | 165ng/μl | 225ng/μl |
Gel analyses and purification.

2. Link aforementioned fragments by overlapping PCR to acquire fragment PcpcB-Kn
| ddwater | 90μl |
| Template1(PcpcB) | 1.2μl(50~100ng/50μl) |
| Template2(Kn) | 1.44μl(50~100ng/50μl) |
| Primer1(10μM) | 4μl |
| Primer2(10μM) | 4μl |
| PrimeSTAR mix | 100μl |
| Total volume | 200μl (50μl*4) |
| (round 1)×8 cycle | 98℃ 3min |
| 98℃ 10s | |
| 53℃ 5s | |
| 72℃ 4s | |
| 72℃ 3min | |
| 12℃ ∞ | |
| (round 2)×35 cycle | 98℃ 3min |
| 98℃ 10s | |
| 63℃ 5s | |
| 72℃ 7s | |
| 72℃ 3min | |
| 12℃ ∞ | |
| Final concentration(after purification) | Sample1+2: 35ng/μlSample3+4: 54ng/μl |
July 9
- Gel analyses for PcpcB-Kn.
- DNA purification to get purified fragment PcpcB-Kn.

July 10
- PCR to get fragment PcpcB-Kn-Down. Gel analyses for PcpcB-Kn-Down.
- Gel extraction to get purified fragment.

July 12
- Link PcpcB-Kn-Down and Up-GFP (had prepared) by overlapping PCR to get the entire sequence of P_dark-GFP and gel analyses.
No correct band is showed. - Transformation of E.coli to amplify pBluescript.
 Transformation
Transformation
Learning the method of transformation, including natural transformation and electro-transformation.
 Testing
Testing
Week 2 (July 13 ~)
 Human Practice
Human Practice
We met Professor Wang and have a long discussion relating to the project feasibility. Because he majors in environment and resource law, we think his questions represent how government and organization will evaluate our project. The plan we had last week can’t cover some of the important problems he raised, so we want to come up with a new plan.
Then, we visited Professor He, an expert working on algae brooms in China. He introduced some of the methods they use to separate cells and also to utilize them. We decided to do some research based on what we learned. Also, we first heard the idea of algae toxin.
 Modeling
Modeling
Discuss the content of modeling
 Construction
Construction
July 13
- Overlapping PCR to get the fragment P_dark-GFP (Annealing temperature changed to a gradience), gel analyses.</br>No correct band is showed.
- PCR for fragment PcpcB-Kn-Down in the way mentioned before to enlarge backup, gel analyses and purification.</br>Final concentration: 244ng/μl
- Inoculation of the transformed E.coli.
July 14
- Overlapping PCR for fragment P_dark-GFP. (Gradient PCR, lengthen annealing and extending time), gel analyses</br>The target band is showed but the concentration is incredibly low and extra bands existed.
- Extraction of plasmid Bluescript, gel analyses.
July 15
- PCR to get the fragment pCPCG2 from genomic DNA of cyanobacteria and Homoup-R1 from plasmid given. Gel analyses.</br>No positive results are shown. It might be the problem of template quantity or quality.
July 16
- PCR to get the fragment pCPCG2 from genomic DNA of cyanobacteria and Homoup-R1 from plasmid given or the previous fragment UP.
pCPCG2 Homoup-R1(from plasmid) Homoup-R1*(from DNA fragment) ddwater 72μl 23μl 22.5μl Template 20μl(70ng/50μl) 0.3μl(1ng/50μl) 0.5μl(<200ng/50μl) Primer1 4μl 2μl 2μl Primer2 4μl 2μl 2μl PrimeSTAR Mix 100μl 50μl 50μl Total volume 200μl(50μl*4) 100μl(50μl*2) 100μl(50μl*2) × 34 cycle 98℃ 3min 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s 98℃ 10s (gradient) 65℃ 5s 65℃ 5s 56℃~62℃ 10s 72℃ 3s 72℃ 3s 72℃ 2s 72℃ 3min 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ 12℃ ∞ Final concentration(after purification) 14.3ng/μl \ 29ng/μl *gradient PCR showed that all annealing temperature can appropriately function.
- Gel analyses for fragment pCPCG2 and Homoup-R1 and gel extraction.

July 17
- PCR for fragments Homoup2, Homoup-R1-R2 and HR(including RBS);
Homoup2 Homoup-R1-R2 HR ddwater 90μl 84μl 92μl Template DNA fragment 4μl(<200ng/50μl) DNA fragment 8μl(116ng/50μl) Synthetic plasmid 0.1μl(1ng/50μl) Primer1 4μl 4μl 4μl Primer2 4μl 4μl 4μl PrimeSTAR Mix 100μl 100μl 100μl Total volume 200μl(50μl*4) 200μl(50μl*4) 200μl(50μl*4) × 34 cycle 98℃ 3min 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s 98℃ 10s 68℃ 5s 65℃ 5s 53℃ 5s 72℃ 3s 72℃ 4s 72℃ 5s 72℃ 3min 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ 12℃ ∞ Final concentration (after purification) 83.5ng/μl 116.4ng/μl 114ng/μl
Gel analyses and purification/gel extraction.

- Overlapping PCR to get the fragments Homoup2-pCPCG2 and P_dark-HR
Homoup-pCPCG2 P_dark-HR ddwater 89μl 92μl Template1 Homoup2 0.3μl(23ng/50μl) Homoup-R1-R2 0.43μl(50ng/50μl) Template2 pCPCG2 3.5μl(50ng/50μl) HR 0.27μl(31ng/50μl) Primer1 4μl 4μl Primer2 4μl 4μl PrimeSTAR Mix 100μl 100μl Total volume 200μl(50μl*4) 200μl(50μl*4) (round 1)×8 cycle 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s 55℃ 5s 45℃ 5s 72℃ 3s 72℃ 5s 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ 12℃ ∞ 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s (gradient) (gradient) 56℃~68℃ 5s 59℃~64℃ 5s 72℃ 3s 72℃ 8s 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ Final concentration (after purification) 121.5ng/μl 187.3ng/μl
*gradient PCR showed that all annealing temperature can appropriately function.</br>Gel analyses and purification/gel extraction.

July 18
- Overlapping PCR for fragment pCPCG2-HR.
*gradient PCR showed that all annealing temperature can appropriately function.</br>Gel analyses and purification/gel extraction.PcpcG2-HR ddwater 89μl Template1 Homoup-PcpcG2 1.6μl(50ng/50μl) Template2 HR 1.4μl(41ng/50μl) Primer1 4μl Primer2 4μl PrimeSTAR Mix 100μl Total volume 200μl(50μl*4) 200μl(50μl*4) 98℃ 3min 98℃ 10s 45.5℃ 5s 72℃ 5s 72℃ 3min 12℃ ∞ 200μl(50μl*4) 98℃ 3min 98℃ 10s (gradient) 59℃~63℃ 5s 72℃ 9s 72℃ 3min 12℃ ∞ Final concentration(after purification) 145.2ng/μl

- Overlapping PCR to get the fragments P_dark-HR-PcpcB and pCPCG2-HR-PcpcB;
Gel analyses and purification/gel extraction.P_dark-HR-PcpcB PcpcG2-HR-PcpcB ddwater 80μl 80μl Template1 P_dark-HR 0.4μl(19ng/50μl) PcpcG2-HR 0.47μl(17ng/50μl) Template2 PcpcB 12μl(50ng/50μl) PcpcB 12μl(50ng/50μl) Primer1 4μl 4μl Primer2 4μl 4μl PrimeSTAR Mix 100μl 100μl Total volume 200μl(50μl*4) 200μl(50μl*4) (round 1)×8 cycle 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s 65℃ 5s 65℃ 5s 72℃ 8s 72℃ 9s 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ (round2) ×33 cycle 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s 57℃ 5s 57℃ 5s 72℃ 12s 72℃ 13s 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ Final concentration (after purification) 165.8ng/μl 170.4ng/μl

- Overlapping PCR to get the whole construction P_dark-HR* and pCPCG2-HR*
Gel analyses and purification/gel extraction.P_dark-HR* PcpcG2-HR* ddwater 88μl 88μl Template1 P_dark-HR-PcpcB 1.1μl(46ng/50μl) PcpcG2-HR-PcpcB 1μl(43ng/50μl) Template2 PcpcB-Kn 2.9μl(50ng/50μl) PcpcB-Kn 2.9μl(50ng/50μl) Primer1 4μl 4μl Primer2 4μl 4μl PrimeSTAR Mix 100μl 100μl Total volume 200μl(25μl*8) 200μl(25μl*8) (round 1)×8 cycle 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s 68℃ 8s 68℃ 8s 72℃ 12s 72℃ 12s 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ (round2) ×33 cycle 98℃ 3min 98℃ 3min 98℃ 10s 98℃ 10s 62℃ 8s 61℃ 8s 72℃ 18s 72℃ 19s 72℃ 3min 72℃ 3min 12℃ ∞ 12℃ ∞ Final concentration (after purification) 8.1ng/μl 14.9ng/μl

The final concentration is not satisfying enough because the extra band existed and gel extraction had to be operated. We adjusted several conditions and tried for a couple of days. During this time, we also changed restriction reaction site and redesigned the primers. See the final version of the protocol of these two fragments at “20150729”.
- PCR for fragments Ecolivk and sll1338 (both are signal sequences) from the synthetic plasmid. But we failed all the time. The main problems are: the concentration of the PCR products are low and for the fragments are too small (both under 120bp) to stay stably on the column when purifying, the purified products could not be used for downstream operation. We tried and adjusted constantly for at least half a month, but no satisfying results were showed.
 Transformation
Transformation
We used plasmid which contains ADS as our positive control to learn the whole process of transformation.
 Testing
Testing
Week 3 (July 20 ~)
 Human Practice
Human Practice
We reorganized the new plan and decided to do our Human Practice project work on the line of the process flow we planned for the desalination, including intake, bio-desalination itself, cell-water separation and the steps after. We can have like a subtopic for each step. In addition, it will be the communication with other iGEM teams and public acceptance.
 Construction
Construction
July 21
- Try different conditions of overlapping PCR for the whole construction of P_dark-HR* and pCPCG2-HR*.
- The final concentration of digestion product was too low for downstream operation. We suspected that it might be misoperation. Also we found out that we used isocaudarners XbaI and BcuI (also called SpeI) as digestion reaction sites, so we changed the two sites later.
July 22
- Try different conditions of overlapping PCR for the whole construction of P_dark-HR* and pCPCG2-HR*.
- Restriction enzyme digestion of pBluescript. CIAP was added in one sample and another one was not. Moreover, the reaction time was lengthened. We found out that CIAP didn’t seem to be that useful in our experiment. But longer reaction time was indeed needed.
July 23
- Try different conditions of overlapping PCR for the whole construction of P_dark-HR* and pCPCG2-HR*.
- We decided to make sure that which method of plasmid‘s digestion reaction is better, single digest or double digest and which enaturation way is better. So we designed several controls using the wrong enzymes as a precursive test. It’s also because that we need to purchase the new enzyme at the time.
Digestion reaction pBluescript(D) pBluescript(S1) pBluescript(S2) P_dark-HR*/ pCPCG2-HR* ddwater 9μl 9μl 9μl Fill up to 30μl 10×(green)buffer 2μl 2μl 2μl 2μl Plasmid/DNA fragment 7.2μl(1μg) 7.2μl(1μg) 7.2μl(1μg) 1μg BcuI 1μl 1μl 1μl 1μl XbaI 1μl 1μl 1μl 1μl Total volume 20μl 20μl 20μl 30μl Reaction 37℃ for 20mins 37℃ for 20mins 37℃ for 20mins Denaturation Gel extraction Purification Thermodynamic denauration
Transformation.Ligation P_dark-HR*(D)/ pCPCG2-HR*(D) P_dark-HR*(S1)/ pCPCG2-HR*(S1) P_dark-HR*(S2)/ pCPCG2-HR*(S2) ddwater Fill up to 10μl Fill up to 10μl Fill up to 10μl 10×buffer 1μl 1μl 1μl Vector DNA 60ng 60ng 60ng Insert DNA 300ng 300ng 300ng Ligase 0.2μl(1WeissU) 0.2μl 0.2μl Total volume 10μl 10μl 10μl Reaction 17℃ for 4h 17℃ for 4h 17℃ for 4h Denaturation Purification/None Purification/None Purification/None
Results:pCPCG2-HR(D) pCPCG2-HR(S1) pCPCG2-HR(S2) Normal Normal Very few colonies P_dark-HR(D) P_dark-HR(S1) P_dark-HR(S2) Normal No colony Very few colonies
 Transformation
Transformation
The natural transformation includes transformation, solid segregation, liquid segregation, the whole process took about three weeks.
 Testing
Testing
Week 4 (July 27 ~)
 Human Practice
Human Practice
We finished some of our work in public acceptance. We found out that there has been data about how short the water is in China, but they are cold charts. Therefore, we made it into data maps so everyone can understand it.
We also did some research in cell-water separation and toxin, especially toxin. There was a few kind of major toxin, but not in our strain gene.
 Modeling
Modeling
Learn how to use modeling tools both mathematical tools like ODEs and PDEs, and software like MATLAB.
 Construction
Construction
July 29
- Overlapping PCR for the whole construction of P_dark-HR* and pCPCG2-HR* using new primers in order to add new restriction enzyme reaction sites XbaI and PstI.
Gel analyses and purification/gel extraction.P_dark-HR* pCPCG2-HR* ddwater 88μl 88μl Template1 P_dark-HR-PcpcB 1.3μl(49.7ng/50μl) pCPCG2-HR-PcpcB 1.4μl(52.8ng/50μl) Template2 PcpcB-Kn 2.9μl(55.7ng/50μl) PcpcB -Kn 2.9μl(55.7ng/50μl) Primer1 4μl 4μl Primer2 4μl 4μl PrimeSTAR Mix 100μl 100μl Total volume 200μl(25μl*8) 200μl(25μl*8) 2-step PCR (round 1)×8 cycle 98℃ 3min 98℃ 10s 72℃ 1min15s 72℃ 3min 12℃ ∞ 2-step PCR (round2) ×33 cycle 98℃ 3min 98℃ 10s 72℃ 1min50s 72℃ 3min 12℃ ∞ Final concentration (after purification) 72.4ng/μl 67.7ng/μl

July 30
- Restriction reaction and ligation reaction of pBluescript and DNA fragments P_dark-HR*, pCPCG2-HR* with XbaI and PstI.
Digestion reaction pBluescript P_dark-HR* pCPCG2-HR* ddwater 45μl 86.8μl 90μl 10×(green) buffer 10μl 8μl 8μl Plasmid/DNA fragment 35μl(1μg) 17.2μl(1μg) 14μl XbaI 5μl 4μl 4μl PstI 5μl 4μl 4μl Total volume 100μl(20μl*5) 120μl(30μl*4) 120μl(30μl*4) Reaction 37℃ for 20min 37℃ for 1h 37℃ for 1h Denaturation Gel extraction Purification Purification
Ligation reaction P_dark-HR*-pBluescript pCPCG2-HR*-pBluescript ddwater 2.8μl 2.8μl 10×buffer 1μl 1μl Vector DNA 1μl(72.1ng) 1μl(72.1ng) Insert DNA 5μl(362ng) 5μl(338.5ng) Ligase 0.3μl 0.3μl Total volume 10μl 10μl Reaction 17℃ for 4h 17℃ for 4h Denaturation Thermodynamic Thermodynamic - Transformation of P_dark-HR*-pBluescript and pCPCG2-HR*-pBluescript.
July 31
- Inoculation: 16 samples for each construction.
Aug. 1
- Bacterial liquid PCR.
Gel analyses.Forward primer 1μl (10μM) Reverse primer 1μl (10μM) Template(bacterial liquid) 1μl 2×Taq Mix 10μl diwater Fill up to 20μl Total volume 20μl ×25 cycle 95℃ 3min 94℃ 15s 68℃ 15s 72℃ 3min50s 72℃ 5min 12℃ ∞

- Plasmid extraction.
- Restriction enzyme reaction examination.

- Transformation of pCPCG2-HR*-pBluescript, using blue-white screen.
- Prepare 10× TBE mother liquor.
- Sequence the examined plasmid containing construction P_dark-HR*-pBluescript.
Aug. 2
- Inoculation: 17 samples.
- Bacterial liquid PCR.

 Transformation
Transformation
The eletro-transformation also includes electrotransformation, solid segregation and liquid segregation, but the whole process took almost four weeks.
We tried the eclctrotransformation of pdark-HR but failed.
 Testing
Testing
Week 5 (August 3 ~)
 Human Practice
Human Practice
We went to Hangzhou Water Treatment Technology Development Center. We had send some of our questions before we arrived about desalination industry and technology. They planned a presentation that is very useful. An important change is instead of thinking replacing those desalination method, we found a new idea to cooperate with them.
 Construction
Construction
Aug. 3
- Plasmid extraction
- Restriction enzyme reaction examination.

Aug. 5
- Propagating cell culture of P_dark-HR*-pBluescript and cell conservation.
Aug. 6
- Plasmid extraction.
 Transformation
Transformation
pdark natural transformation
In the noon of Aug. 5
Inoculated wildtype cyanobacteria into the flask and make sure the initial OD was about 0.1;
In the evening of Aug. 7
When the OD of our culture reached 0.4-0.6, collect the cell and mix the DNA, cyanobacteria and liquid medium, meanwhile set a negative control culture (without DNA), put them into illumination incubator at 30℃;
Aug. 7
Shook by hand for about 4 times from morning(every 2-3 hours), after 8 hours, took 200ul culture of each tube, spread them on different solid plates which contained 20 micrograms kanamycin per milliliter, put the plate into illumination incubator at 30℃.
pcpcG2 electro-transformation
At the noon of Aug. 8
We inoculated wildtype cyanobacteria into the flask and make sure the initial OD was about 0.1; prepare the electric tubes
 Testing
Testing
Week 6 (August 10 ~)
 Human Practice
Human Practice
We brought some of our work in public acceptance to Shanghai Science and Technology Museum. This presentation made us want to do it more prepared to a bigger audience.
 Modeling
Modeling
Reviewing literature and get ready for modeling
 Construction
Construction
Aug. 10
- Prepare 34mg/ml chloramphenicol.
- Propagating cell culture of pCPCG2-HR*-pBluescript and cell conservation.
Aug. 11
- Plasmid extraction.
Aug. 12
- Transformation of BBa_J04450.
Aug. 13
- Inoculation: 10 samples.
Aug. 14
- Plasmid extraction.
- Linearize plasmid by restriction enzyme reaction. Extract the linearized plasmid.
Aug. 16
- Inoculation of DH5α for component cell making.
 Transformation
Transformation
pcpcG2 electro-transformation
In the evening of Aug. 10
Put electric tubes into 60℃ dryer for at least 2 hours, sterilized them by UV before electrotransformation.
Collect the cyanobacteria cell and diluted the cell with sterilized ddH2O washed it for 5 times,then diluted it with sterile ddH2O, mixed with pcpcg2-HR plasmid, transfered them into the electric tube, tunk for several times so that the liquid can reach to the bottom, installed the tube into switch, did electro-transformation, then transfer the mixture to BG11 culture
In the evening of Aug. 11
Centrifuged the cyanobacteria, diluted the cell with 400ul liquid culture, mixed with 5ml soft agar BG11 medium, spread on solid medium which contained 20 micrograms kanamycin per milliliter, put the plate into illumination incubator at 30 ℃.
 Testing
Testing
Week 7 (August 17 ~)
 Human Practice
Human Practice
We went to Shanghai Water Supply Authority; it is a supplement visit for something we write. It is good to verify what you write to reality.
 Construction
Construction
Aug. 17
- Component cell making.
Aug. 20
- Producted linearized Pdark-GFP, amplified GFP fragments from constructed plasmid GFP.
- Agarose gel electrophoresis for PCR products.
- Extracted positive plasmids (16 tubes).
- Inoculated 14 D3 colonies and negative control C2/D2 for 2 tubes. (16 tubes in total)
Aug. 21
- Extracted plasmids from the 16 tubes inoculated on 8.20.
- Agarose gel electrophoresis test and restriction enzyme digestion test for plasmids above.(20μl system included 1μl SacI and 1μl EcoRI.)
- Colony PCR test for D3 and C2/D2.
- Prepared homologous recombination system (as is shown below).
- Transformation and plating afterwards.
| GFP-1* | GFP-2* | GFP-3a | GFP-3b | |
|---|---|---|---|---|
| ddwater | 12μl | 11μl | 10μl | 9μl |
| 5x CEII Buffer | 4μl | 4μl | 4μl | 4μl |
| Linearized plasmid Pdark-HR(-GFP-)-1 | 3μl | 3μl | 3μl | 3μl |
| GFP fragment | 1μl | 0μl | 1μl | 2μl |
| Exnase II | 0μl( | 2μl | 2μl | 2μl |
- *GFP-1&2 are 2 control groups.
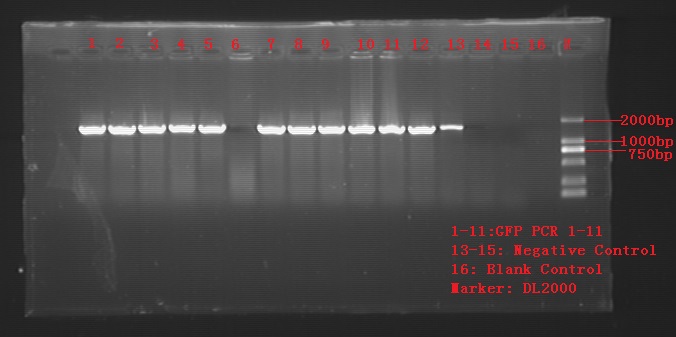
Aug. 22
Colony PCR for recombination cells. 12tubes for single colonies of GFP 3a and 3b, 3 tubes for control groups GFP-1 and GFP-2.
| GFP-3a/3b/1/2 | |
|---|---|
| ddwater | 144μl |
| Template | 0.5μl(each tube) |
| Primer M13F(10μM) | 8μl |
| Primer GFP-R(10μM) | 8μl |
| 2xTaq Mix | 160μl |
| Total volume | 320μl(20μl*16) |
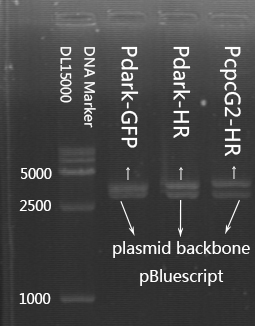
As is shown in the figure, product no.6 showed negative and control no.1 showed positive. Most of the products qualified the length of 1400bp or so.
Aug. 23
Mass prep of Pdark-GFP plasmid (No.1,4).
 Transformation
Transformation
pdark natural transformation
In the evening of Aug 18
We picked 4 colonies (marked with 1,2,3 & 4) from the pdark-HR plate and spread them on another BG11 plate in parallel seprately to do solid segregation
In the evening of Aug. 20
We scraped the grown cell from the plate, which is 1,3 & 4 to do colony PCR, 1st liquid segregation and solid storage
pre-experiment of pdark-HR
In the evening of Aug. 20
Inoculated 16 wildtype & 16 pdark-HR (use pdark-HR from electro-transformation), added 11g/L or 22g/L NaCl & retinene at the beginning
In the evening of Aug. 22
Induced with darkness, took the first 4 samples at 9pm
In the evening of Aug. 23
Cultivated with white light, took the second 4 samples at 9pm.
 Testing
Testing
Week 8 (August 24 ~)
 Human Practice
Human Practice
We struggled to come up with the report of intake. We planned to do collaboration, yet, we want something that will cover the necessity of this project. At last, we decide to add a section to explain the trend of water supply and the estimated trend of desalination resource water. This will provide us a stronger why.
 Modeling
Modeling
Determine the main idea about modeling
 Construction
Construction
Aug. 25
PCR cloning of fragment Pdark-GFP and gel extraction. Final concentration: 119.5ng/μl (tube1); 304ng/μl (tube2)
Aug. 26
Linearization of pSB1C3 plasmid by PCR.
| BBa_J0450 | |
|---|---|
| ddwater | 160μl |
| Template | 32μl |
| Primer pSB-F(10μM) | 8μl |
| Primer pSB-R(10μM) | 8μl |
| PrimeSTAR Mix | 200μl |
| Total volume | 400μl(50μl*8) |
DpnI digestion for 2h. Gel extraction.
Shipping part Pdark-GFP, Pdark-HR and PcpcG2-HR into pSB1C3 plasmid via PCR - Homologous recombination method (24 tubes).
| Parts | Control-1(*3) | Control-2(*3) | |
|---|---|---|---|
| ddwater | 300μl | 14μl | 15μl |
| 5x CEII Buffer | 100μl | 4μl | 4μl |
| Linearized plasmid pSB1C3 | 25μl | 1μl | 0μl |
| Fragment (Pdark-GFP/HR,PcpcG2-HR) | 25μl | 1μl | 1μl |
| Exnase II | 50μl | 0μl | 2μl |
Agarose Gel electrophoresis test.
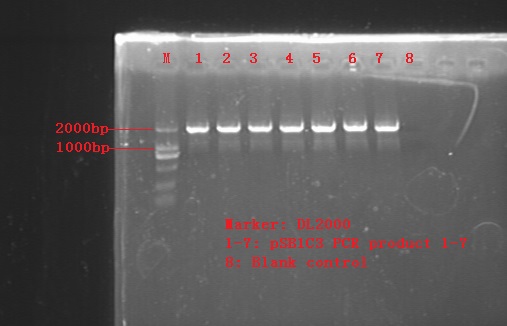
Aug. 27
Transformation into DH5a (4 tubes not including no-fragment control) and plating afterwards.
* (Lack of control group led to a failure of this transformation.)
Test with colony PCR and restriction enzyme reaction (XbaI and PstI).
Plasmid extraction (Pdark-GFP).
Aug. 28
Linearization of official vector plasmid pSB1C3.
| BBa_J0450 | |
|---|---|
| ddwater | 160μl |
| Template | 32μl |
| Primer pSB-F(10μM) | 8μl |
| Primer pSB-R(10μM) | 8μl |
| PrimeSTAR Mix | 200μl |
| Total volume | 400μl(50μl*8) |
DpnI digestion for 2h. Gel extraction.
PCR for fragment PcpcG2-HR.
| PcpcG2-HR | Pdark-HR | |
|---|---|---|
| ddwater | 220μl | 220μl |
| Template | 10μl | 10μl |
| Primer UHKD-F(10μM) | 10μl | 10μl |
| Primer UHKD-R(10μM) | 10μl | 10μl |
| PrimeSTAR Mix | 250μl | 250μl |
| Total volume | 500μl(50μl*8) | 500μl(50μl*8) |
Agarose gel electrophoresis test and gel extraction for Pdark-HR and PcpcG2-HR fragment.
Shipping part Pdark-HR and PcpcG2-HR into pSB1C3 plasmid via PCR - Homologous recombination method.
Second try of homologous recombination.
| Pdark-HR | PcpcG2-HR | Non-fragment control | |
|---|---|---|---|
| ddwater | 12μl | 12μl | 13μl |
| 5x CEII Buffer | 4μl | 4μl | 4μl |
| Linearized plasmid pSB1C3 (80ng/μl) | 1μl | 1μl | 1μl |
| Fragment | 1μl | 1μl | 0μl |
| Exnase II | 2μl | 2μl | 2μl |
Transformation into DH5a (3 tubes) and plating afterwards.
Spread plates and cultured for 14h in 37℃.
Aug. 29
Inoculation of Pdark-GFP, Pdark-HR and PcpcG2-HR transformed DH5a. (11 tubes each, 3 from control group)
Aug. 30
Bacteria Liquid PCR fpr Pdark-GFP, Pdark-HR and PcpcG2-HR. Agarose gel electrophoresis test.
Pdark-GFP:
| pSB-vF | 1μl (10μM) |
| GFP-R | 1μl (10μM) |
| Bacterial liquid(Pdark-GFP) | 1μl |
| 2×Taq Mix | 10μl |
| ddwater | Fill up to 20μl |
| Total volume | 20μl |
| 95℃ 3min | |
| 94℃ 15s | ×28 cycle |
| 48℃ 15s | |
| 72℃ 1min20s | |
| 72℃ 5min | |
| 12℃ ∞ |
Pdark-HR:
| pSB-vF | 1μl (10μM) |
| SS-HR-R | 1μl (10μM) |
| Bacterial liquid(Pdark-GFP) | 1μl |
| 2×Taq Mix | 10μl |
| ddwater | Fill up to 20μl |
| Total volume | 20μl |
| 95℃ 3min | |
| 94℃ 15s | ×28 cycle |
| 48℃ 15s | |
| 72℃ 50s | |
| 72℃ 5min | |
| 12℃ ∞ |
PcpcG2-HR:
| pSB-vF | 1μl (10μM) |
| SS-HR-R | 1μl (10μM) |
| Bacterial liquid(Pdark-GFP) | 1μl |
| 2×Taq Mix | 10μl |
| ddwater | Fill up to 20μl |
| Total volume | 20μl |
| 95℃ 3min | |
| 94℃ 15s | ×28 cycle |
| 48℃ 15s | |
| 72℃ 60s | |
| 72℃ 5min | |
| 12℃ ∞ |
 Transformation
Transformation
pdark natural transformation
In the evening of Aug. 29
2nd liquid segregation of pdark-HR
pcpcG2 electro-transformation
In the evening of Aug. 24
We picked 4 colonies (marked with 1,2,3 & 4) from the pcpcG2-HR plate and spread them on another BG11 plate in parallel seprately to do solid segregation
In the evening of Aug. 30
We scraped the grown cell from the plate, which is 2,3 & 4 to do colony PCR, 1st liquid segregation and solid storage
pre-experiment of pdark-HR
Aug. 24
Took the third 4 samples at 9am & fourth 4 samples at 9pm.
Aug. 25
Took the fifth 4 samples at 9am & sixth 4 samples at 9pm.
Aug. 26
Took the seventh 4 samples at 9am & eighth 4 samples at 9pm.
pdark-GFP electro-transformation
In the evening of Aug. 24
Set culture.
In the evening of Aug. 26
Electro-transformation of pdark-GFP.
In the evening of Aug. 27
Collected half of the cell and mixed with 5ml soft agar, then poured on solid medium plate.
 Testing
Testing
Week 9 (August 31 ~)
 Human Practice
Human Practice
We worked on the desalination part. Most of the information in this section we already knew, so, it is an easy week.
 Modeling
Modeling
Construct the dynamic system model using ODE
 Construction
Construction
Aug. 31
- Plasmid extraction.
- Double restriction enzyme digestion test for Pdark-GFP, Pdark-HR and PcpcG2-HR.
| Restriction Enzyme Digestion | |
|---|---|
| ddwater | 360μl |
| FD buffer | 450μl |
| XbaI | 45μl |
| PstI | 45μl |
| Template | 1μl/tube |
| Total volume | 900μl(20μl*39) |
Agarose gel extraction test.
Sep. 2
Gel anlysis of the restriction enzyme digestion product for a second time.
Sep. 3
Sequencing samples. Pdark-GFP-3, Pdark-GFP-8, PcpcG2-HR-4,PcpcG2-HR-6, Pdark-HR-2, Pdark-HR-4.
Sep. 6
All sequenced sample were successful and right. Site-directed mutation on PcpcG2-HR to eliminate the EcoRI sites via PCR method.
| PcpcG2-HR-Mut | |
| ddwater | 105μl |
| G2Mut-F | 10μl (10μM) |
| G2Mut-R | 10μl (10μM) |
| PcpcG2-HR(pSB1C3) | 2μl |
| PrimeStar Mix | 125μl |
| Total volume | 250μl(25μl*8)( |
| 98℃ 3min | |
| 98℃ 10s | ×32 cycle |
| 65℃ 5s | |
| 72℃ 30s | |
| 72℃ 5min | |
| 12℃ ∞ |
DpnI digestion for 2h. Electrophoresis test and gel extraction afterwards.
Homologous recombination of PcpcG2-HR-Mut.
| PG2-Mut | Control-1 | |
|---|---|---|
| ddwater | 12μl | 14μl |
| 5x CEII Buffer | 4μl | 4μl |
| PCR product | 2μl | 2μl |
| Exnase II | 2μl | 0μl |
37℃ for 30min. Product storaged in -20℃ overnight.
 Transformation
Transformation
pdark natural transformation
In the evening of Aug. 31
3rd liquid segregation of pdark-HR
In the evening of Sep. 3
4th liquid segregation of pdark-HR
In the evening of Sep. 6
5th liquid segregation of pdark-HR
pcpcG2 electro-transformation
In the evening of Sep. 2
2nd liquid segregation of pcpcG2-HR
In the evening of Sep. 4
3rd liquid segregation of pcpcG2-HR
In the evening of Sep. 6
4th liquid segregation of pcpcG2-HR
 Testing
Testing
second desalination test of pdark-HR
Sep. 2
inoculated 24 wildtype & 24 pdark-HR (use pdark-HR from colony 3) at 9pm
Sep. 4
induced with darkness at 9pm, added 22g/L NaCl & retinene, then took the first 6 samples (3 parallel samples for each)
Sep. 5
cultivated with white light at 9am, then took the second 6 samples,
took the third 6 samples at 10am
took the fourth 6 samples at 11am
took the fifth 6 samples at 1pm
took the sixth 6 samples at 9pm
Sep. 6
took the seventh 6 samples at 9am
took the eighth 6 samples at 9pm
Week 10 (September 7 ~)
 Human Practice
Human Practice
Connecting everything together.
 Modeling
Modeling
Coding and simulating using MATLAB and plot the result
 Construction
Construction
Sep. 7
Transformation of PcpcG2-HR-Mut into DH5a and plating. After incubation for 12h, Inoculated single colonies into liquid LB culture (10 tubes for test group and 2 for control group).
Sep. 8
Plasmid extraction.
Double restriction enzyme digestion.
| Restriction Enzyme Digestion | |
|---|---|
| ddwater | 280μl |
| FD buffer | 40μl |
| XbaI | 20μl |
| EcoRI | 20μl |
| Template | 2μl/tube |
| Total volume | 400μl(20μl*20) |
- Template included PcpcG2-HR-Mut, PcpcG2-HR and 0 control.
Agarose gel electrophoresis test.
Sep. 9
Sequencing PcpcG2-HR-Mut-4 and PcpcG2-HR-Mut-9.
Sep. 10
Sequencing results proved right.
Sep. 17
Functional fragments restriction enzyme digestion.
| Pdark-GFP | Pdark-HR | PcpcG2-HR | |
|---|---|---|---|
| ddwater | 15μl | 15μl | 15μl |
| FD buffer | 2μl | 2μl | 2μl |
| XbaI | 1μl | 1μl | 1μl |
| PstI | 1μl | 1μl | 1μl |
| Template | 1.3μl | 1μl | 0.9μl |
| Total volume | 20μl | 20μl | 20μl |
 Transformation
Transformation
pdark natural transformation
In the evening of Sep. 11
6th liquid segregation of pdark-HR
pcpcG2 electro-transformation
In the evening of Sep. 11
5th liquid segregation of pcpcG2-HR
pdark-GFP electro-transformation
Sep. 8
We picked 1 colony (only 1 colony appeared) from the pdark-GFP plate and spread them on another BG11 plate in line to do solid segregation
 Testing
Testing
first desalination test of pcpcG2-HR
Sep. 7
Inoculated 15 wildtype & 15 pcpcG2-HR (use pcpcG2-HR from colony 2) at 10pm.
Sep. 9
Induced with green light at 10pm, added 22g/L NaCl & retinene, then took the first 6 samples (3 parallel samples for each).
Sep. 10
Cultivated with red light at 10am, then took the second 6 samples,
Took the third 6 samples at 2pm.
Took the fourth 6 samples at 10pm.
Sep. 11
Took the fifth 6 samples at 10am.
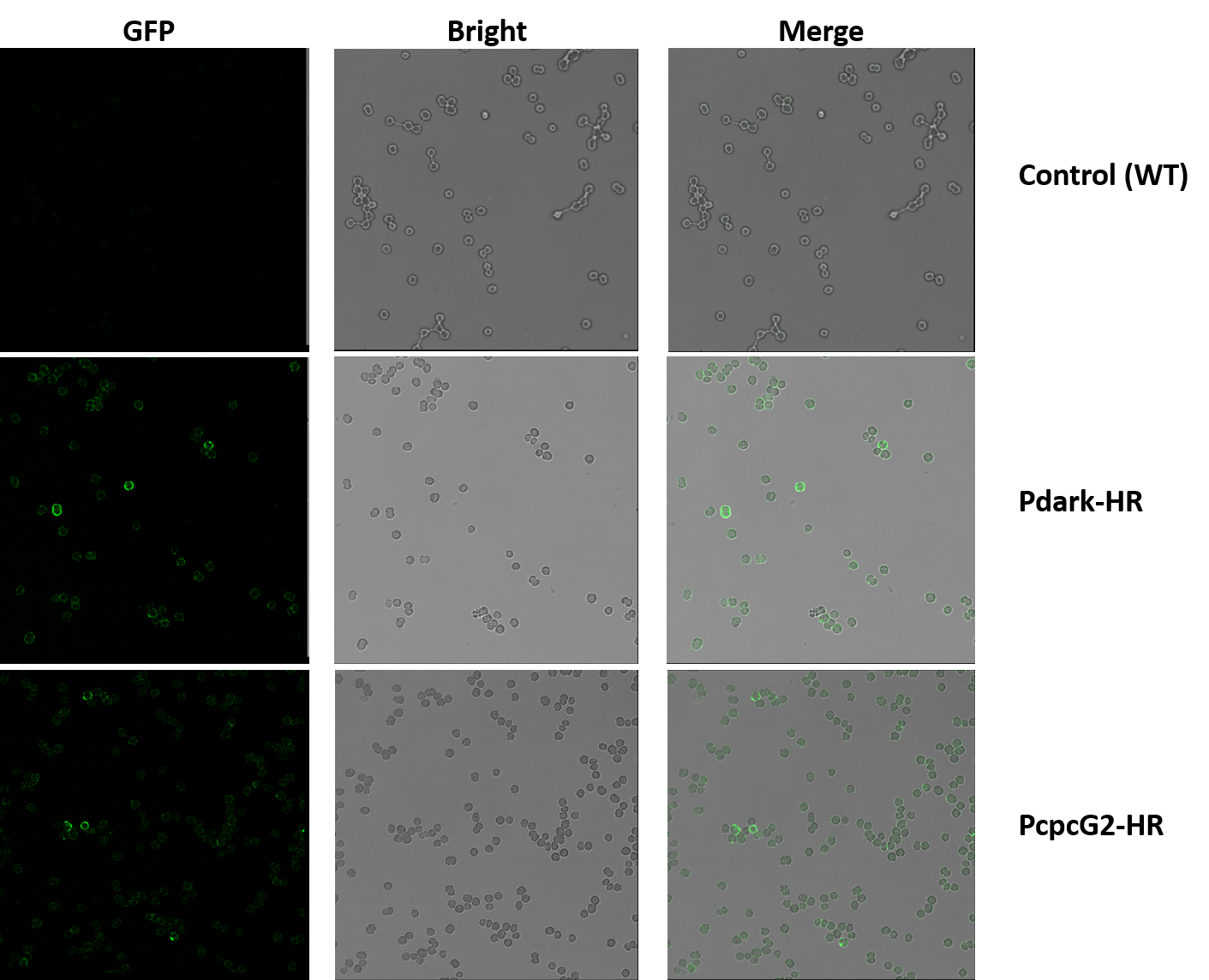
Detection of Membrane Localization of Halorhodopsin by Immunocytochemistry
To examine whether the HR expressed successfully onto the cell membrane, we designed an immunocytochemical experiment. For that the C terminal of the HR sequence we designed contains a His-tag, we used anti-His mAb as the primary antibody and goat anti-mouse IgG Alexa Fluor 488(green fluorescence) as the second antibody. And if a loop of green fluorescence can be seen under the confocal microscope, we will ensure that the HR expressed successfully onto the cell membrane. A few things should be paid attention to:
- The incubating condition can be overnight at 4℃ or 2 hours at 37℃. But if a better result is wanted, use the former condition.
- Be careful that the procedures involving fluorescent antibodies need to be operated under lucifugal condition.
Sep. 15
1ml cyanobacteria liquid for each sample was prepared, including four experiment groups--Pdark-HR(green), Pdark-HR(red), PcpcG2-HR(green) and PcpcG2-HR(red), which had been cultured to OD730=0.6 and induced for 12h, and controls—wildtype(green) and wildtype(red), which were not been induced.
Anti-His mAb was applied with PBS in proportion of 1:100 as the primary antibody. After 2 hours’ incubating(at 37℃) and washing steps, goat anti-mouse IgG Alexa Fluor 488(green fluorescence) and goat anti-mouse IgG Alexa Fluor 594(red fluorescence) were applied respectively with PBS in proportion of 1:1000 as the second antibodies.
After 2 hours’ incubating(at 37℃) and washing steps, the cyanobacteria pellets were resuspended with 50μl PBS. Six slides were prepared. While observing under a confocal microscope, we found out that the cyanobacteria has spontaneous red fluorescence. Several positive results can be seen but the signal is not strong enough. So we decided to optimize the protocol and redo it.
Sep.15 evening to Sep. 16
1ml cyanobacteria liquid for each sample was prepared, including four experiment groups--Pdark-HR(1:100), Pdark-HR(1:50), PcpcG2-HR(1:100) and PcpcG2-HR(1:50), which had been cultured to OD730=0.6 and induced for 16h, and controls—wildtype(1:100) and wildtype(1:50), which had been induced for 16h as well.
The primary antibody anti-His mAb was applied with PBS in proportion of 1:100 or 1:50 as mentioned. The samples were incubated at 4℃ overnight. The second antibody goat anti-mouse IgG Alexa Fluor 488(green fluorescence) was applied with PBS in proportion of 1:300 and the samples were incubated at 37℃ for 2 hours.
After washing the samples, the cyanobacteria pellets were resuspended with 20μl PBS. Six slides were prepared. The results were better than the former one. Concerning of time, we only got the micrographs of the samples containing the primary antibody with the concentration of 1:50.
In the experiment groups (Pdark-HR and PcpcG2-HR), many loops of green fluorescence can be seen under the confocal microscope while the control groups (wildtype) cannot, which indicates that the halorhodopsin expressed successfully on the cell membrane of the genetic cyanobacteria instead of any other position in the cells.