Team:Tsinghua/Basic Part
Basic Parts
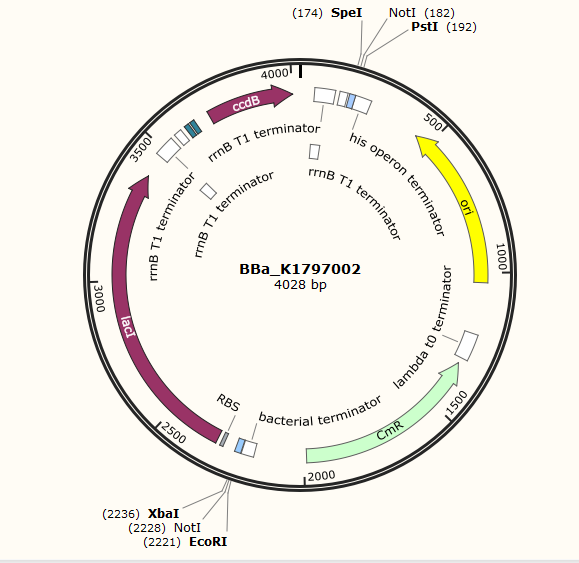
Part:BBa_K1797002
IPTG-inducible CcdB
This part includes lacI and lac operator which are intended for the control of the expression of CcdB. LacI is constitutively expressed and binds to lac operator to block the expression of CcdB. In the presence of IPTG, lacI is released from operator and CcdB can be expressed. CcdB is a poisonous protein for E.coli since it affects the activity of DNA gyrase and inhibits the segregation of chromosomal DNA. Thus, when IPTG is added,the bacteria containing this part will go through cell death.
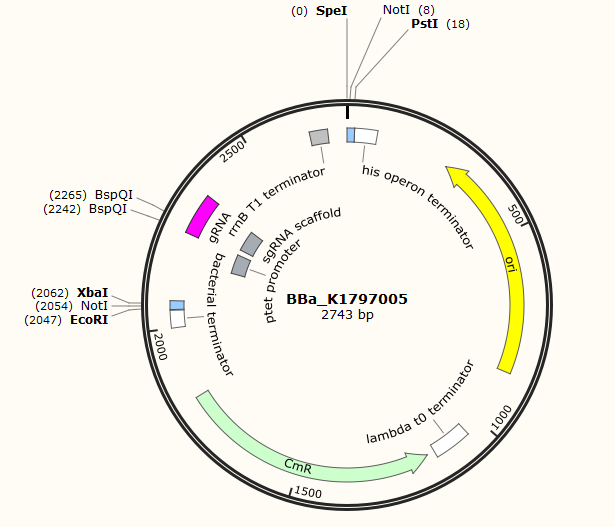
Part:BBa_K1797005
sgRNA
This part includes sgRNA, which can be used to do site-specific gene editing when CRISPR/Cas9 system is present. The sgRNA changeable region can be changed due to the purpose of uses. Users can use BspQI to digest the part and ligate with a DNA sequence that has BspQI compatible ends. The DNA sequence is designed according to the gene sequence that needs gene editing.
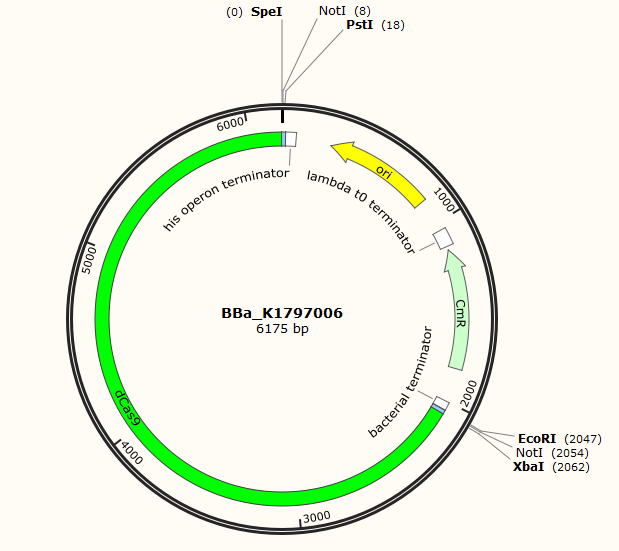
Part:BBa_K1797006
dCas9
CRISPR Cas9 is a very popular gene editing system. This part includes an engineered dCas9 which is still able to bind to the DNA under the guidance of gRNA. However, it loses its catalytic activity. Actually, dCas9 has a very promising future. For example, We can use dCas9 to block the recruitment of transcription machine.
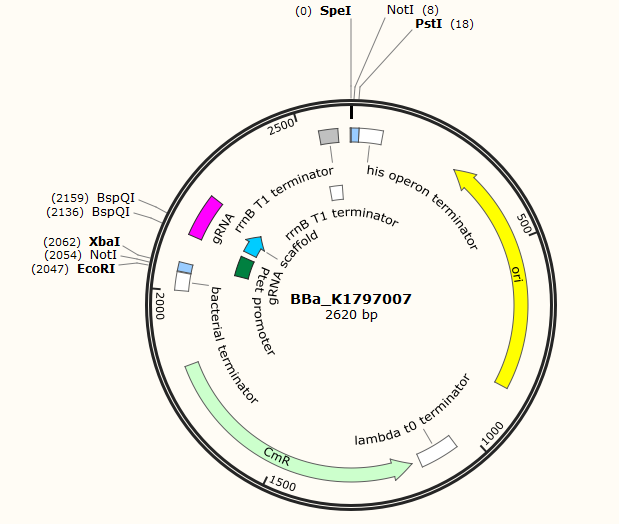
Part:BBa_K1797007
Tet promoter-sgRNA-Terminator
This part is modified from BBa_K1797005. We added promoter and terminator before and after the sgRNA for the convenience of users. Users can change the sequence of sgRNA(except for the sgRNA scaffold) for their own purpose.
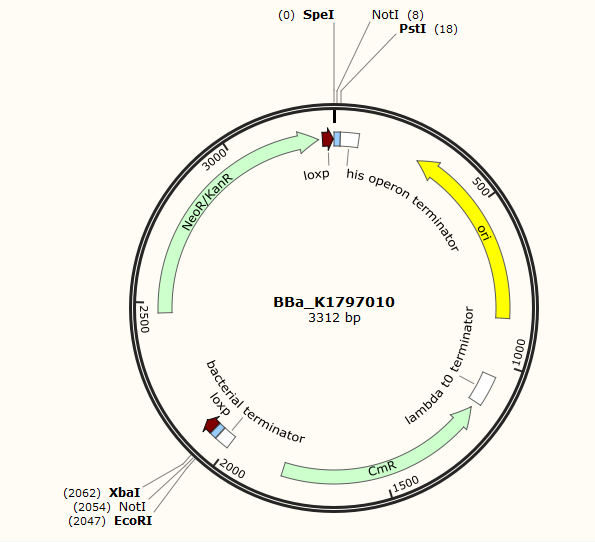
Part:BBa_K1797010
Kana cassette flanked by loxP site
This part includes a translational kanamycin resistance gene(in prokaryotes) or a neomycin resistance gene(in eukaryotes) and flanking loxP sites, which can be recognized by Cre recombinase. We can use DNA recombination technology to knock this part into E.coli genome and the antibiotic resistance gene helps us find the right colonies. Then if Cre is expressed in the bacteria, this kana cassette can be excised and a knock out experiment is completed.
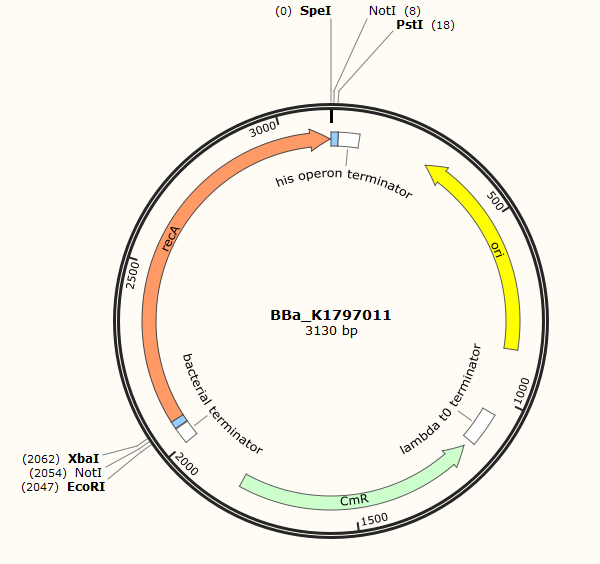
Part:BBa_K1797011
RecA
RecA is an essential protein for DNA repair and maintenance. Researchers (Stahl F W, Stahl M M, Malone R E. Red-mediated recombination of phage lambda in a recA− recB− host[J]. Molecular and General Genetics MGG, 1978, 159(2): 207-211.) have found that red recombination system works better in recA+ strains. To maximize the efficiency of red recombination system, we need to introduce recA exogenously in plasmid form though recA is intrinsically existent in E.coli. To have a clearer view of red recombination system, you can refer to BBa_K1797014 and BBa_K1797013.
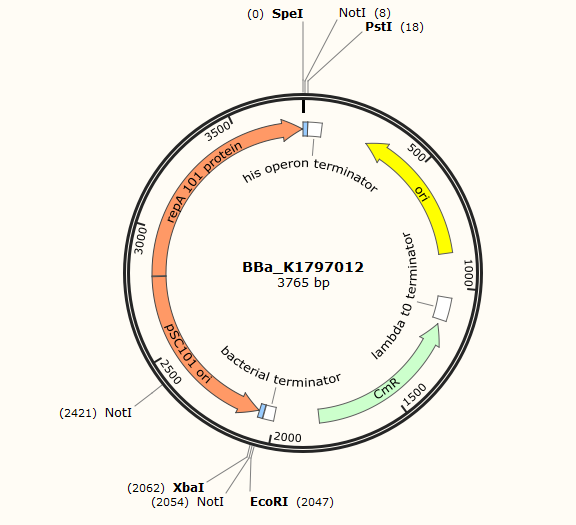
Part:BBa_K1797012
pSC101 Ori-a temperature sensitive replication origin
pSC101 ori is a low-copy replication origin that requires the Rep101 protein, which is a temperature-sensitive protein. When bacteria containing a plasmid with this ori is cultured at 37℃, the plasmid will be lost.
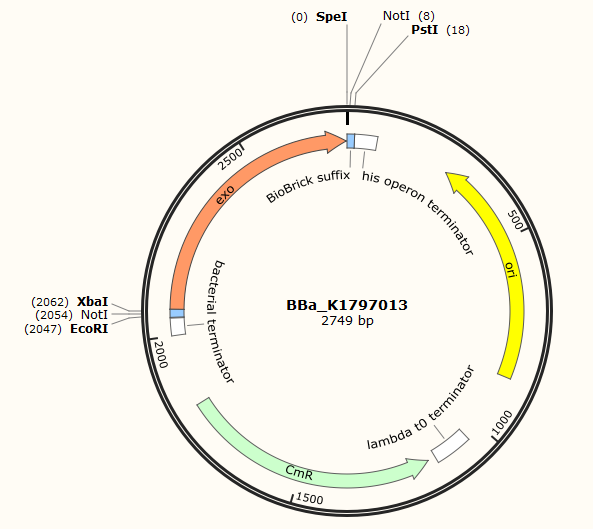
Part:BBa_K1797013
Exo-an exonuclease in the Red homologous recombination system
Red recombination system is a efficient homologous recombination system found in lamda phage. It includes three main proteins, gam, bet and exo. Gam is able to bind RecBCD, inhibiting its ability to degrade exogenous linear DNA. Bet binds to the 3' overhang produced by exo and protects it, preventing its degradation by nucleases. Red recombination system is becoming popular in terms of gene editing. The common procedure is to PCR a certain fragment(often a kana cassette for easy screening later) flanked with homologous regions(about 20-50bp) first. Then the linear fragment and the red recombination system is introduced into bacteria for homologous recombination.This part contains one component of red recombination system-exo. Exo is an exonuclease which can bite a dsDNA and form a 3' overhang.
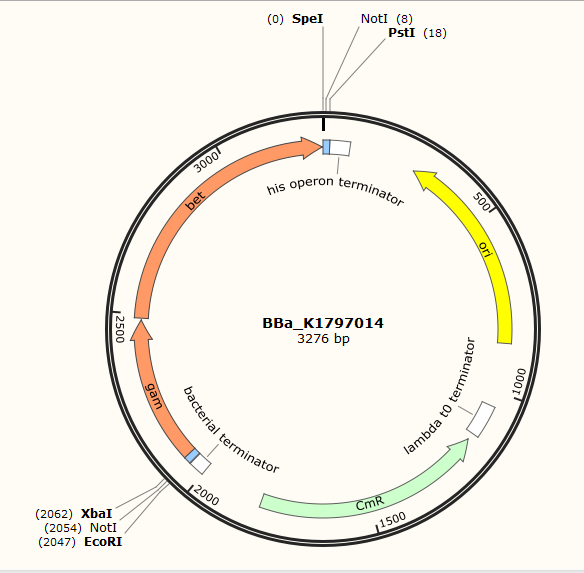
Part:BBa_K1797014
Gam and Bet-Two Components of the Red Recombination System
Red recombination system is a efficient homologous recombination system found in lamda phage. It includes three main proteins, gam, bet and exo. Gam is able to bind RecBCD, inhibiting its ability to degrade exogenous linear DNA. Bet binds to the 3' overhang produced by exo and protects it, preventing its degradation by nucleases. Red recombination system is becoming popular in terms of gene editing. The common procedure is to PCR a certain fragment(often a kana cassette for easy screening later) flanked with homologous regions(about 20-50bp) first. Then the linear fragment and the red recombination system is introduced into bacteria for homologous recombination.This part contains two components-gam and bet.
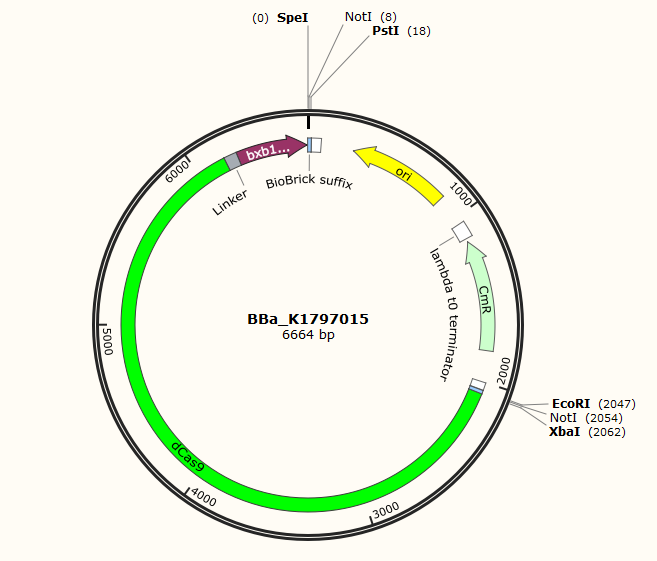
Part:BBa_K1797015
dCas9-Long Linker (Gly-Gly-Ser-Gly)x3-Bxb1 gp35
In this part, we fuse dCas9(BBa_K1797006) and catalytic domain of Bxb1 gp35(BBa_K907000) with a long linker(BBa_K243006). Bxb1 gp35 is a site-specific recombinase. Due to its specific recognition site, the use of recombinase is limited. To solve this problem, we fuse its catalytic domain with dCas9. Then where the Bxb1 gp35 is going to function is decided by gRNA.
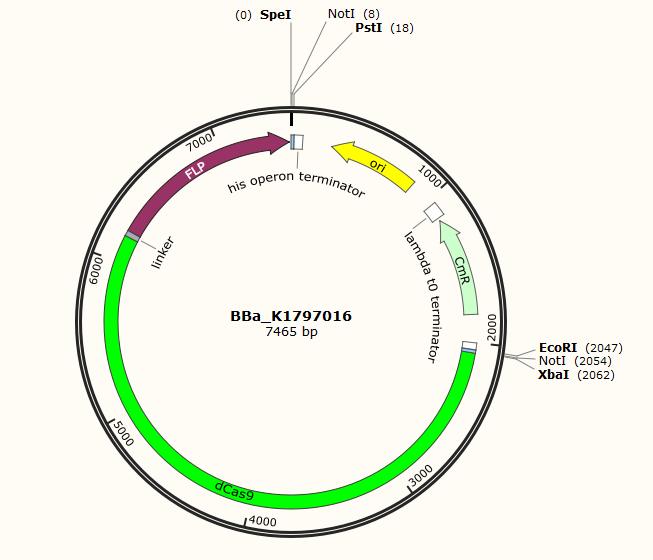
Part:BBa_K1797016
dCas9-Long Linker (Gly-Gly-Ser-Gly)x3-Flp
In this part, we fuse dCas9(BBa_K1797006) and catalytic domain of Flp(BBa_K1015001) with a long linker(BBa_K243006). Flp is a site-specific recombinase. Due to its specific recognition site, the use of recombinase is limited. To solve this problem, we fuse its catalytic domain with dCas9. Then where the Flp is going to function is decided by gRNA.
</p> </div>