Difference between revisions of "Team:Waterloo/Modeling/CaMV Biology"
m |
|||
| Line 5: | Line 5: | ||
<h1>Cauliflower Mosaic Virus (CaMV) Biology</h1> | <h1>Cauliflower Mosaic Virus (CaMV) Biology</h1> | ||
| − | <p> | + | <p>Cauliflower Mosaic Virus (CaMV) is a member of the caulimovirus genus of pararetroviruses. CaMV is an ideal virus for the study of CRISPR plant defense for several reason. <ol> |
| − | + | <li>Most importantly, CaMV only infects plants, making it significantly safer to work with</li>. | |
| + | <li>Second, it can infect <em>Arabidopsis thaliana</em>, the plant model organism for genetics, meaning many of the tools for genetic engineering already work.</li> | ||
| + | <li>Finally, the CaMV genome is double stranded for much of the virus lifecycle, which means it can be easily targeted by CRISPR-Cas9 for cutting.</li> | ||
| + | </ol> | ||
| + | </p> | ||
<h2> Stuff from Viral Replication Page </h2> | <h2> Stuff from Viral Replication Page </h2> | ||
<div class="panel-group" id="bio background" role="tablist" aria-multiselectable="true"> | <div class="panel-group" id="bio background" role="tablist" aria-multiselectable="true"> | ||
| + | <div class="panel panel-default"> | ||
| + | <div class="panel-heading" role="tab" id="lifeCycle"> | ||
| + | <h3 class="panel-title"> | ||
| + | <a class="collapsed" role="button" data-toggle="collapse" data-parent="#bio background" href="#collapseCycle" aria-expanded="false" aria-controls="collapseCycle"> | ||
| + | Overview of Life Cycle | ||
| + | <span class="hidden-xs toggle-arrow pull-right">Panel Toggle</span> | ||
| + | </a> | ||
| + | </h3> | ||
| + | </div> | ||
| + | <div id="collapseCycle" class="panel-collapse collapse" role="tabpanel" aria-labelledby="lifeCycle"> | ||
| + | <div class="panel-body"> | ||
| + | <p>The lifecycle of a pararetrovirus is rather similar to that of a retrovirus such as HIV. Both types of viruses use a reverse transcriptase to turn viral mRNA into DNA. However, unlike retroviruses, pararetroviruses do not integrate into the host genome. Instead, thousands of minichromosomes of viral genome are imported into the nucleus, where they are transcribed by host machinery<cite ref="Hohn2013"></cite>.</p> | ||
| + | <p>Here we present a brief overview of the CaMV replication cycle, compiled from Khelifa et al. 2010 (<cite ref="Khelifa2010"></cite>), Yasaka et al. 2014 (<cite ref="Yasaka2014"></cite>), Hohn, Fütterera & Hull 1997 (<cite ref="Hohn1997"></cite>), and Citovsky, Knorr, & Zambryski 1991 (<cite ref="Citovsky1991"></cite>)</p> | ||
| + | <ol> | ||
| + | <li>The virion enters the host cell, moves to the nuclear pore, and the viral genome is imported in the nucleus</li> | ||
| + | <li>Discontinuities in the genomes are repaired to form minichromosomes</li> | ||
| + | <li>Minichromosomes are used as templates to produce 35S RNA, 19S RNA, and 8S RNA</li> | ||
| + | <li>P6 is translated from 19S RNA and accumulates in electron-dense inclusion bodies (edIBs)</li> | ||
| + | <li>P6 transactivates production of P1-P5 from 35S RNA</li> | ||
| + | <li>Packaging occurs. The exact process is complicated, but either <cite ref="Hohn1997"></cite>:</li> | ||
| + | <ul> | ||
| + | <li>35S RNA is packaged into virions along with P5 then is reverse transcribed within the capsid, or</li> | ||
| + | <li>35S RNA is reverse transcribed in the edIBs before packaging</li> | ||
| + | </ul> | ||
| + | <li>The virion may either reinfect the nucleus to increase the copy number of the minichromosome (early in the infection) or it may leave the cell</li> | ||
| + | </ol> | ||
| + | <p>Due to the complicated nature of viral packaging, we decided in our model to combine the packaging reaction into one term, dependent on P4, P5, and unspliced 35S RNA concentration.</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
<div class="panel panel-default"> | <div class="panel panel-default"> | ||
<div class="panel-heading" role="tab" id="genomeandStuff"> | <div class="panel-heading" role="tab" id="genomeandStuff"> | ||
| Line 24: | Line 58: | ||
<h4>Overview of Genome</h4> | <h4>Overview of Genome</h4> | ||
<p> | <p> | ||
| − | The CaMV genome consists of circular, double-stranded DNA approximately 8000 bp in length <cite ref="Franck1980"></cite>. The genome contains three discontinuities typical of pararetroviruses <cite ref="Franck1980"></cite> <cite ref="Hohn2013"></cite>. Discontinuities are repaired within the nucleus to form covalently closed circular ( | + | The CaMV genome consists of circular, double-stranded DNA approximately 8000 bp in length <cite ref="Franck1980"></cite>. The genome contains three discontinuities typical of pararetroviruses <cite ref="Franck1980"></cite> <cite ref="Hohn2013"></cite>. Discontinuities are repaired within the nucleus to form covalently closed circular DNA (cccDNA) <cite ref="Khelifa2010"></cite>. There are seven open reading frames (ORFs) which code for six proteins of known function; P1-P6 (labeled gp2-gp7 in the diagram below) <cite ref="Khelifa2010"></cite> <cite ref="Hohn1997"></cite>. Two main RNAs are produced from cccDNA: pregenomic 35S RNA and subgenomic 19S RNA, as well as the small 8S RNA <cite ref="Guilley1982"></cite>. |
</p> | </p> | ||
<figure> | <figure> | ||
| Line 37: | Line 71: | ||
<h4>Pregenomic 35S RNA</h4> | <h4>Pregenomic 35S RNA</h4> | ||
<p> | <p> | ||
| − | 35S RNA is a complete transcript of the CaMV genome with additional 180 nt terminal repeats <cite ref="Guilley1982"></cite> <cite ref="Hohn1985"></cite>. It has two primary functions. The first is to act as an intermediate in genome replication. That is, it's reverse transcribed either in a viral factory or within the capsid to recover gapped dsDNA <cite ref="Pfeiffer1983"></cite>. It's second function is as polycistronic mRNA coding for P1-P5. | + | 35S RNA is a complete transcript of the CaMV genome with additional 180 nt terminal repeats <cite ref="Guilley1982"></cite> <cite ref="Hohn1985"></cite>. It has two primary functions. The first is to act as an intermediate in genome replication. That is, it's reverse transcribed either in a viral factory or within the capsid to recover gapped dsDNA <cite ref="Pfeiffer1983"></cite>. It's second function is as polycistronic mRNA coding for P1-P5. Translation of P1-P5 from the 35S promoter must be transactivated by P6 <cite ref="Futterer1991"></cite>. 35S RNA also undergoes alternative splicing to regulate P1 and P2, which are potentially toxic in high enough concentrations <cite ref="Kiss-Laszlo1995"></cite> <cite ref="Froissart2004"></cite>. |
</p> | </p> | ||
<h4>Subgenomic 19S RNA</h4> | <h4>Subgenomic 19S RNA</h4> | ||
<p> | <p> | ||
| − | The 19S RNA is a 2.3 kb <cite ref="Covey1981"></cite> transcript which contains the ORF for P6 <cite ref="Khelifa2010"></cite>. P6 is constitutively produced from 19S RNA and it's many functions are described in it's own section | + | The 19S RNA is a 2.3 kb <cite ref="Covey1981"></cite> transcript which contains the ORF for P6 <cite ref="Khelifa2010"></cite>. P6 is constitutively produced from 19S RNA and it's many functions are described in it's own section. |
</p> | </p> | ||
<h4>8S RNA</h4> | <h4>8S RNA</h4> | ||
<p> | <p> | ||
| − | 8S RNA is 610-680 nt long <cite ref="Olszewski1983"></cite> and is transcribed from the leader sequence of 35S RNA. It's believed that it serves as a decoy for RNA interference, which normally targets the 35S leader <cite ref="Blevins2011"></cite> <cite ref="Hohn2015"></cite> | + | 8S RNA is 610-680 nt long <cite ref="Olszewski1983"></cite> and is transcribed from the leader sequence of 35S RNA. It's believed that it serves as a decoy for RNA interference, which normally targets the 35S leader <cite ref="Blevins2011"></cite> <cite ref="Hohn2015"></cite>. |
</p> | </p> | ||
<h4>Movement Protein (P1)</h4> | <h4>Movement Protein (P1)</h4> | ||
<p> | <p> | ||
| − | The movement protein (MP) of CaMV facilitates intercellular spread of virions <cite ref="Lucas2006"></cite>. P1 binds to pectin methyltranferase <cite ref="Chen2000"></cite> and ultimately forms intercellular tubules though which virions are transported to neighbouring cells <cite ref="Kastell1996"></cite>. This protein may also bind cooperatively to RNA to direct 35S and/or 19S RNA to neighbouring cells <cite ref="Citovsky1990"></cite>, which suggests the protein-RNA complex may also move between cells without a capsid | + | The movement protein (MP) of CaMV facilitates intercellular spread of virions <cite ref="Lucas2006"></cite>. P1 binds to pectin methyltranferase, a cell wall associated protein <cite ref="Chen2000"></cite>, and ultimately forms intercellular tubules though which virions are transported to neighbouring cells <cite ref="Kastell1996"></cite>. This protein may also bind cooperatively to RNA to direct 35S and/or 19S RNA to neighbouring cells <cite ref="Citovsky1990"></cite>, which suggests the protein-RNA complex may also move between cells without a capsid. |
</p> | </p> | ||
<h4>Aphid Transmission Factor (P2)</h4> | <h4>Aphid Transmission Factor (P2)</h4> | ||
<p> | <p> | ||
| − | The aphid transmission factor (ATF) is a protein which allows for host-to-host transmission of the virus using aphids as vectors. P2 binds to | + | The aphid transmission factor (ATF) is a protein which allows for host-to-host transmission of the virus using aphids as vectors. P2 binds to the aphid stylets and P3, which allows virions to associate with the stylet upon feeding <cite ref="Drucker2002"></cite>. Its primary function is host-to-host transmission, but it does produce one electron-lucent inclusion body (elIB) which has minor effects on the system. To form the elIB, P2 associates with microtubules and are transported along the cytoskeleton to a central location. In this process some P3 associates with the P2 and is transported to the elIB <cite ref="Martiniere2009"></cite>. Finally, a small percent (~5%) of virions find their way to the elIB <cite ref="Bak2013"></cite>. |
</p> | </p> | ||
<h4>Virion-Associated Protein (P3)</h4> | <h4>Virion-Associated Protein (P3)</h4> | ||
<p> | <p> | ||
| − | The virion associated protein (VAP) is a 15 kDa protein is weakly associated with the viral capsid <cite ref="Mesnard1994"></cite>. It's implicated in both host-to-host <cite ref="Drucker2002"></cite> and cell-to-cell <cite ref="Kobayashi2002"></cite> transmission | + | The virion associated protein (VAP) is a 15 kDa protein is weakly associated with the viral capsid <cite ref="Mesnard1994"></cite>. It's implicated in both host-to-host <cite ref="Drucker2002"></cite> and cell-to-cell <cite ref="Kobayashi2002"></cite> transmission. |
</p> | </p> | ||
<h4>Capsid Protein (P4)</h4> | <h4>Capsid Protein (P4)</h4> | ||
<p> | <p> | ||
| − | The capsid protein (CP) is a precursor to numerous other polypeptides which we refer to as P4 subunits. The P4 subunits include P37, P44, P40, P80, P90, and P96, with P80 and P96 being homodimers composed of P37 and P44 respectively <cite ref="MartinezIzquierdo1987"></cite>. | + | The capsid protein (CP) is a precursor to numerous other polypeptides which we refer to as P4 subunits. The P4 subunits include P37, P44, P40, P80, P90, and P96, with P80 and P96 being homodimers composed of P37 and P44 respectively <cite ref="MartinezIzquierdo1987"></cite>. A viral capsid is formed from 420 subunits composed into 3 layers<cite ref="Cheng1992"></cite>. |
</p> | </p> | ||
<h4>Reverse Transcriptase (P5)</h4> | <h4>Reverse Transcriptase (P5)</h4> | ||
<p> | <p> | ||
| − | The | + | The P6 protein reverse transcribes 35S RNA to form partially double stranded DNA (pdsDNA) <cite ref="Pfeiffer1983"></cite>. The pdsDNA is then encapsidated by P4 subunits to produce virions. It's unknown if reverse transcription occurs within the capsid or in the viroplasm <cite ref="Hohn1997"></cite>. There is evidence that in Hepatitis B (a similar pararetrovirus) that RT first binds to the pregenome, then the RT-RNA complex is recognized by CP to form a capsid, after which reverse transcription finally occurs <cite ref="Ganem1994"></cite>. |
</p> | </p> | ||
<h4>Transactivator/Viroplasmin Protein (P6)</h4> | <h4>Transactivator/Viroplasmin Protein (P6)</h4> | ||
<p> | <p> | ||
| − | The transactivator/viroplasmin protein (TAV), as it's name suggests, is a protein primarily involved in transactivating the five other proteins <cite ref="Futterer1991"></cite> and condenses into viroplasms <cite ref="Kobayashi2003"></cite>. The viroplasms, aka viral factories or electron-dense inclusion bodies (edIBs), sequester 35S RNA transcripts and produces the other proteins required for viral spread and assembly <cite ref="Mazzolini1989"></cite>. The purpose is two-fold: | + | The transactivator/viroplasmin protein (TAV), as it's name suggests, is a protein primarily involved in transactivating the five other proteins <cite ref="Futterer1991"></cite> and condenses into viroplasms <cite ref="Kobayashi2003"></cite>. The viroplasms, aka viral factories or electron-dense inclusion bodies (edIBs), sequester 35S RNA transcripts and produces the other proteins required for viral spread and assembly <cite ref="Mazzolini1989"></cite>. The purpose is two-fold: concentrating the nucleic acid and protein in a smaller volume to increase reaction rates, and protecting the replication machinery from cell defenses <cite ref="Mazzolini1989"></cite>. |
</p> | </p> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<div class="panel panel-default"> | <div class="panel panel-default"> | ||
<div class="panel-heading" role="tab" id="rnaInterference"> | <div class="panel-heading" role="tab" id="rnaInterference"> | ||
Revision as of 05:03, 17 September 2015
Cauliflower Mosaic Virus (CaMV) Biology
Cauliflower Mosaic Virus (CaMV) is a member of the caulimovirus genus of pararetroviruses. CaMV is an ideal virus for the study of CRISPR plant defense for several reason.
- Most importantly, CaMV only infects plants, making it significantly safer to work with .
- Second, it can infect Arabidopsis thaliana, the plant model organism for genetics, meaning many of the tools for genetic engineering already work.
- Finally, the CaMV genome is double stranded for much of the virus lifecycle, which means it can be easily targeted by CRISPR-Cas9 for cutting.
Stuff from Viral Replication Page
The lifecycle of a pararetrovirus is rather similar to that of a retrovirus such as HIV. Both types of viruses use a reverse transcriptase to turn viral mRNA into DNA. However, unlike retroviruses, pararetroviruses do not integrate into the host genome. Instead, thousands of minichromosomes of viral genome are imported into the nucleus, where they are transcribed by host machinery.
Here we present a brief overview of the CaMV replication cycle, compiled from Khelifa et al. 2010 (), Yasaka et al. 2014 (), Hohn, Fütterera & Hull 1997 (), and Citovsky, Knorr, & Zambryski 1991 ()
- The virion enters the host cell, moves to the nuclear pore, and the viral genome is imported in the nucleus
- Discontinuities in the genomes are repaired to form minichromosomes
- Minichromosomes are used as templates to produce 35S RNA, 19S RNA, and 8S RNA
- P6 is translated from 19S RNA and accumulates in electron-dense inclusion bodies (edIBs)
- P6 transactivates production of P1-P5 from 35S RNA
- Packaging occurs. The exact process is complicated, but either :
- 35S RNA is packaged into virions along with P5 then is reverse transcribed within the capsid, or
- 35S RNA is reverse transcribed in the edIBs before packaging
- The virion may either reinfect the nucleus to increase the copy number of the minichromosome (early in the infection) or it may leave the cell
Due to the complicated nature of viral packaging, we decided in our model to combine the packaging reaction into one term, dependent on P4, P5, and unspliced 35S RNA concentration.
Overview of Genome
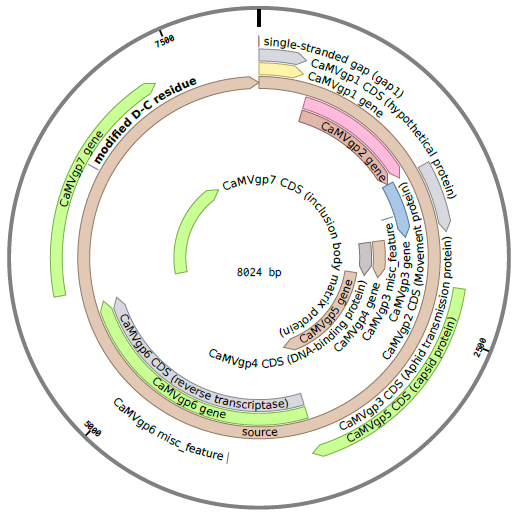
The CaMV genome consists of circular, double-stranded DNA approximately 8000 bp in length . The genome contains three discontinuities typical of pararetroviruses . Discontinuities are repaired within the nucleus to form covalently closed circular DNA (cccDNA) . There are seven open reading frames (ORFs) which code for six proteins of known function; P1-P6 (labeled gp2-gp7 in the diagram below) . Two main RNAs are produced from cccDNA: pregenomic 35S RNA and subgenomic 19S RNA, as well as the small 8S RNA .
Pregenomic 35S RNA
35S RNA is a complete transcript of the CaMV genome with additional 180 nt terminal repeats . It has two primary functions. The first is to act as an intermediate in genome replication. That is, it's reverse transcribed either in a viral factory or within the capsid to recover gapped dsDNA . It's second function is as polycistronic mRNA coding for P1-P5. Translation of P1-P5 from the 35S promoter must be transactivated by P6 . 35S RNA also undergoes alternative splicing to regulate P1 and P2, which are potentially toxic in high enough concentrations .
Subgenomic 19S RNA
The 19S RNA is a 2.3 kb transcript which contains the ORF for P6 . P6 is constitutively produced from 19S RNA and it's many functions are described in it's own section.
8S RNA
8S RNA is 610-680 nt long and is transcribed from the leader sequence of 35S RNA. It's believed that it serves as a decoy for RNA interference, which normally targets the 35S leader .
Movement Protein (P1)
The movement protein (MP) of CaMV facilitates intercellular spread of virions . P1 binds to pectin methyltranferase, a cell wall associated protein , and ultimately forms intercellular tubules though which virions are transported to neighbouring cells . This protein may also bind cooperatively to RNA to direct 35S and/or 19S RNA to neighbouring cells , which suggests the protein-RNA complex may also move between cells without a capsid.
Aphid Transmission Factor (P2)
The aphid transmission factor (ATF) is a protein which allows for host-to-host transmission of the virus using aphids as vectors. P2 binds to the aphid stylets and P3, which allows virions to associate with the stylet upon feeding . Its primary function is host-to-host transmission, but it does produce one electron-lucent inclusion body (elIB) which has minor effects on the system. To form the elIB, P2 associates with microtubules and are transported along the cytoskeleton to a central location. In this process some P3 associates with the P2 and is transported to the elIB . Finally, a small percent (~5%) of virions find their way to the elIB .
Virion-Associated Protein (P3)
The virion associated protein (VAP) is a 15 kDa protein is weakly associated with the viral capsid . It's implicated in both host-to-host and cell-to-cell transmission.
Capsid Protein (P4)
The capsid protein (CP) is a precursor to numerous other polypeptides which we refer to as P4 subunits. The P4 subunits include P37, P44, P40, P80, P90, and P96, with P80 and P96 being homodimers composed of P37 and P44 respectively . A viral capsid is formed from 420 subunits composed into 3 layers.
Reverse Transcriptase (P5)
The P6 protein reverse transcribes 35S RNA to form partially double stranded DNA (pdsDNA) . The pdsDNA is then encapsidated by P4 subunits to produce virions. It's unknown if reverse transcription occurs within the capsid or in the viroplasm . There is evidence that in Hepatitis B (a similar pararetrovirus) that RT first binds to the pregenome, then the RT-RNA complex is recognized by CP to form a capsid, after which reverse transcription finally occurs .
Transactivator/Viroplasmin Protein (P6)
The transactivator/viroplasmin protein (TAV), as it's name suggests, is a protein primarily involved in transactivating the five other proteins and condenses into viroplasms . The viroplasms, aka viral factories or electron-dense inclusion bodies (edIBs), sequester 35S RNA transcripts and produces the other proteins required for viral spread and assembly . The purpose is two-fold: concentrating the nucleic acid and protein in a smaller volume to increase reaction rates, and protecting the replication machinery from cell defenses .
Someone please update, maybe Laura?
Lifecycle Overview
Infection
Replication
Intercellular Spread
CaMV Genome Structure
Natural Plant Defenses
e.g. RNAi